山大集中講義2019.03.08;Day 2
Day 2
- 始めるにあたって,プロジェクトフォルダを置くこと
- 下のコードを実行して,必要なパッケージを読み込んでおくこと
library(tidyverse)
# マカーの呪文
old = theme_set(theme_gray(base_family = "HiraKakuProN-W3"))
library(brms)## Loading required package: Rcpp## Loading 'brms' package (version 2.7.0). Useful instructions
## can be found by typing help('brms'). A more detailed introduction
## to the package is available through vignette('brms_overview').
## Run theme_set(theme_default()) to use the default bayesplot theme.線形モデル1;回帰分析
データセットの確認
read_csv('baseball2019.csv') %>%
dplyr::select(height,weight) %>%
na.omit() %>%
ggplot(aes(x=weight,y=height))+geom_point()## Parsed with column specification:
## cols(
## .default = col_double(),
## Name = col_character(),
## team = col_character(),
## position = col_character(),
## bloodType = col_character(),
## throw.by = col_character(),
## batting.by = col_character(),
## birth.place = col_character(),
## birth.day = col_date(format = ""),
## 背番号 = col_character()
## )## See spec(...) for full column specifications.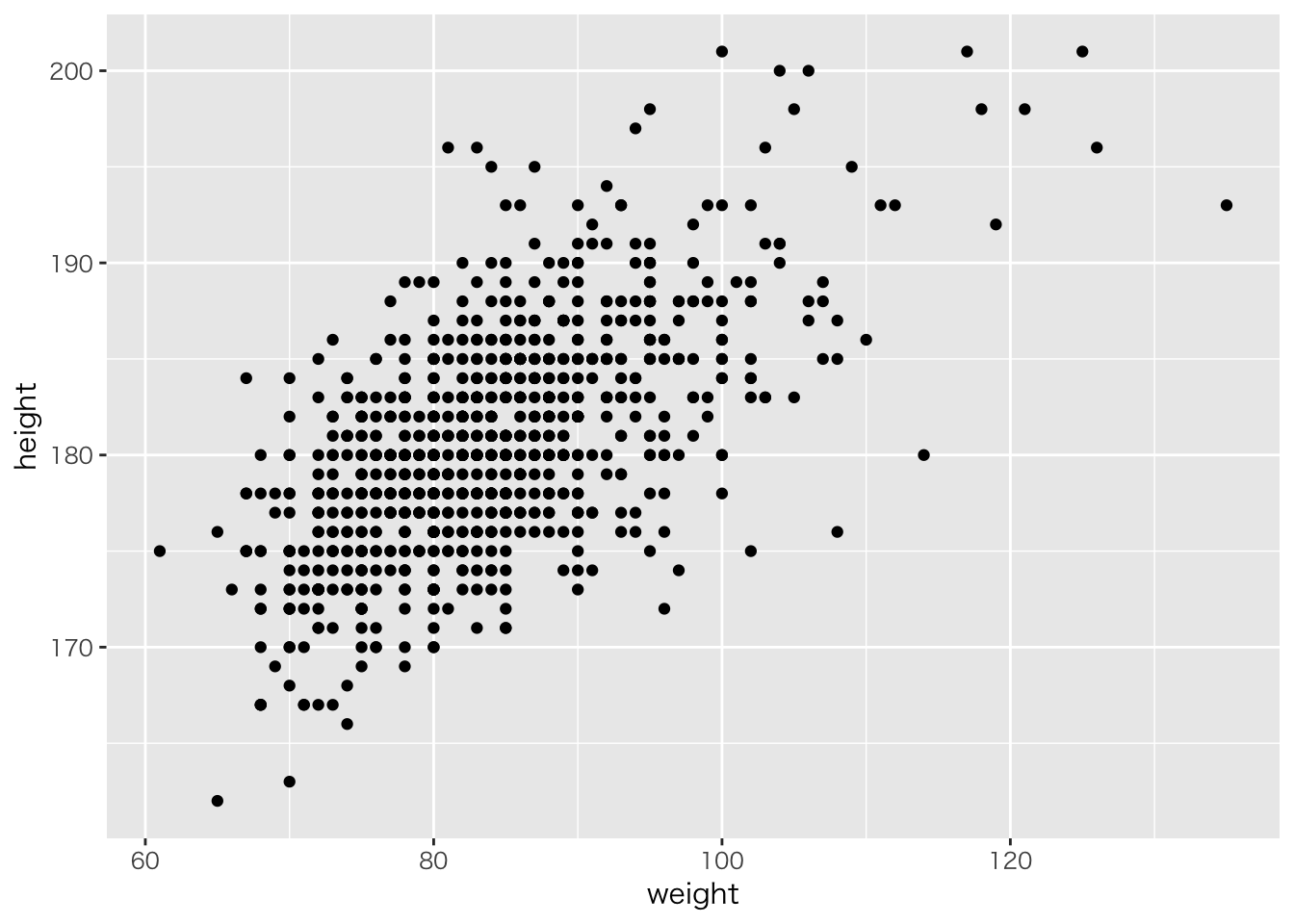
read_csv('baseball2019.csv') %>%
dplyr::select(height,weight) %>%
na.omit() %>%
ggplot(aes(x=weight,y=height))+geom_point()+geom_smooth(method='lm',se=FALSE)## Parsed with column specification:
## cols(
## .default = col_double(),
## Name = col_character(),
## team = col_character(),
## position = col_character(),
## bloodType = col_character(),
## throw.by = col_character(),
## batting.by = col_character(),
## birth.place = col_character(),
## birth.day = col_date(format = ""),
## 背番号 = col_character()
## )## See spec(...) for full column specifications.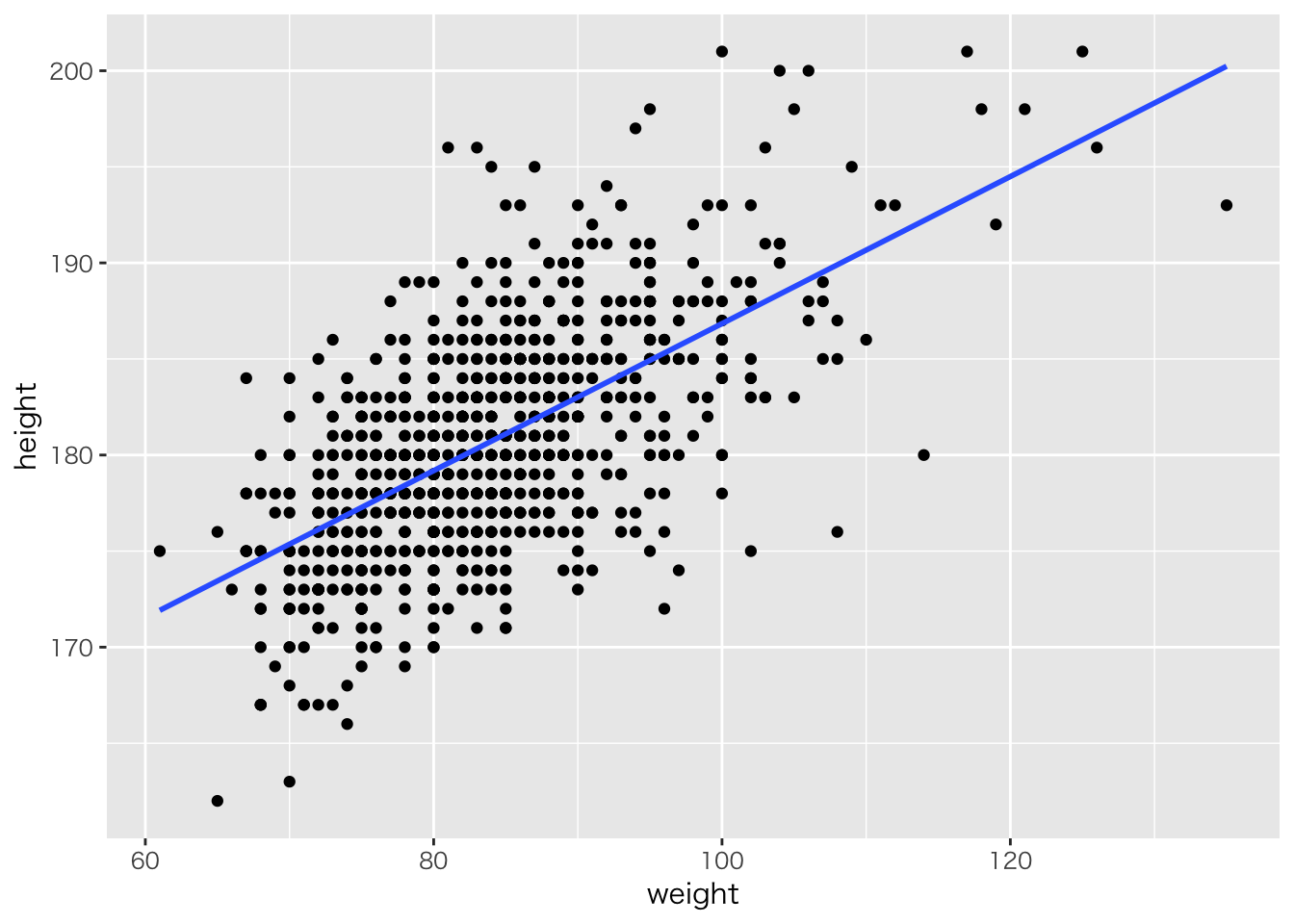
推定方法1;最尤推定
read_csv('baseball2019.csv') %>%
dplyr::select(height,weight) %>%
na.omit() -> baseball_dat## Parsed with column specification:
## cols(
## .default = col_double(),
## Name = col_character(),
## team = col_character(),
## position = col_character(),
## bloodType = col_character(),
## throw.by = col_character(),
## batting.by = col_character(),
## birth.place = col_character(),
## birth.day = col_date(format = ""),
## 背番号 = col_character()
## )## See spec(...) for full column specifications.result.lm <- lm(height~weight,data=baseball_dat)
summary(result.lm)##
## Call:
## lm(formula = height ~ weight, data = baseball_dat)
##
## Residuals:
## Min 1Q Median 3Q Max
## -13.9016 -2.8402 -0.0261 2.6653 16.4306
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 148.57283 1.33809 111.03 <2e-16 ***
## weight 0.38267 0.01586 24.13 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 4.37 on 916 degrees of freedom
## Multiple R-squared: 0.3886, Adjusted R-squared: 0.388
## F-statistic: 582.3 on 1 and 916 DF, p-value: < 2.2e-16オブジェクトの中身を見てみる
result.lm$coefficients## (Intercept) weight
## 148.5728253 0.3826734result.lm$residuals %>% head()## 1 2 3 4 5 6
## -5.7426354 -10.1000627 -5.3347159 1.4306308 0.7519172 -6.0134296result.lm$fitted.values %>% head()## 1 2 3 4 5 6
## 175.7426 181.1001 180.3347 179.5694 182.2481 183.0134重回帰分析
read_csv('baseball2019.csv') %>%
dplyr::select(height,weight,years) %>%
na.omit() -> baseball_dat2## Parsed with column specification:
## cols(
## .default = col_double(),
## Name = col_character(),
## team = col_character(),
## position = col_character(),
## bloodType = col_character(),
## throw.by = col_character(),
## batting.by = col_character(),
## birth.place = col_character(),
## birth.day = col_date(format = ""),
## 背番号 = col_character()
## )## See spec(...) for full column specifications.result.lm2 <- lm(height~weight+years,baseball_dat2)
summary(result.lm2)##
## Call:
## lm(formula = height ~ weight + years, data = baseball_dat2)
##
## Residuals:
## Min 1Q Median 3Q Max
## -14.0395 -2.7728 0.0038 2.7152 15.7869
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 149.19556 1.33278 111.943 < 2e-16 ***
## weight 0.38466 0.01571 24.483 < 2e-16 ***
## years -0.13985 0.03207 -4.361 1.44e-05 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 4.328 on 915 degrees of freedom
## Multiple R-squared: 0.4011, Adjusted R-squared: 0.3998
## F-statistic: 306.4 on 2 and 915 DF, p-value: < 2.2e-16標準化偏回帰係数
データを標準化する。
baseball_dat2 %>% scale %>% as.data.frame -> baseball_dat2z
baseball_dat2z %>% head()| height | weight | years |
|---|---|---|
| -1.9106909 | -1.4158734 | 0.0798896 |
| -1.7316733 | 0.1225781 | -0.1443876 |
| -1.0156030 | -0.0972006 | 1.2012756 |
| 0.0585025 | -0.3169794 | 1.2012756 |
| 0.4165376 | 0.4522463 | 0.7527212 |
| -0.6575678 | 0.6720251 | 1.2012756 |
baseball_dat2z %>% summary()## height weight years
## Min. :-3.3428 Min. :-2.5148 Min. :-1.0415
## 1st Qu.:-0.6576 1st Qu.:-0.6466 1st Qu.:-0.8172
## Median :-0.1205 Median :-0.0972 Median :-0.3687
## Mean : 0.0000 Mean : 0.0000 Mean : 0.0000
## 3rd Qu.: 0.5956 3rd Qu.: 0.4522 3rd Qu.: 0.5284
## Max. : 3.6389 Max. : 5.6170 Max. : 4.3412標準化するということがどういうことか,確認しておこう。
result.lm3 <- lm(height~weight+years,data=baseball_dat2z)
summary(result.lm3)##
## Call:
## lm(formula = height ~ weight + years, data = baseball_dat2z)
##
## Residuals:
## Min 1Q Median 3Q Max
## -2.51331 -0.49637 0.00068 0.48607 2.82613
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.325e-15 2.557e-02 0.000 1
## weight 6.266e-01 2.560e-02 24.483 < 2e-16 ***
## years -1.116e-01 2.560e-02 -4.361 1.44e-05 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.7747 on 915 degrees of freedom
## Multiple R-squared: 0.4011, Adjusted R-squared: 0.3998
## F-statistic: 306.4 on 2 and 915 DF, p-value: < 2.2e-16線形モデル2;(重)回帰分析の特徴
残差の平均値
result.lm$residuals %>% mean## [1] 1.512317e-16説明変数と残差の共分散
cov(baseball_dat$weight,result.lm$residuals)## [1] -2.617788e-14data.frame(pred=baseball_dat$weight,resid=result.lm$residuals) %>%
ggplot(aes(x=pred,y=resid))+geom_point()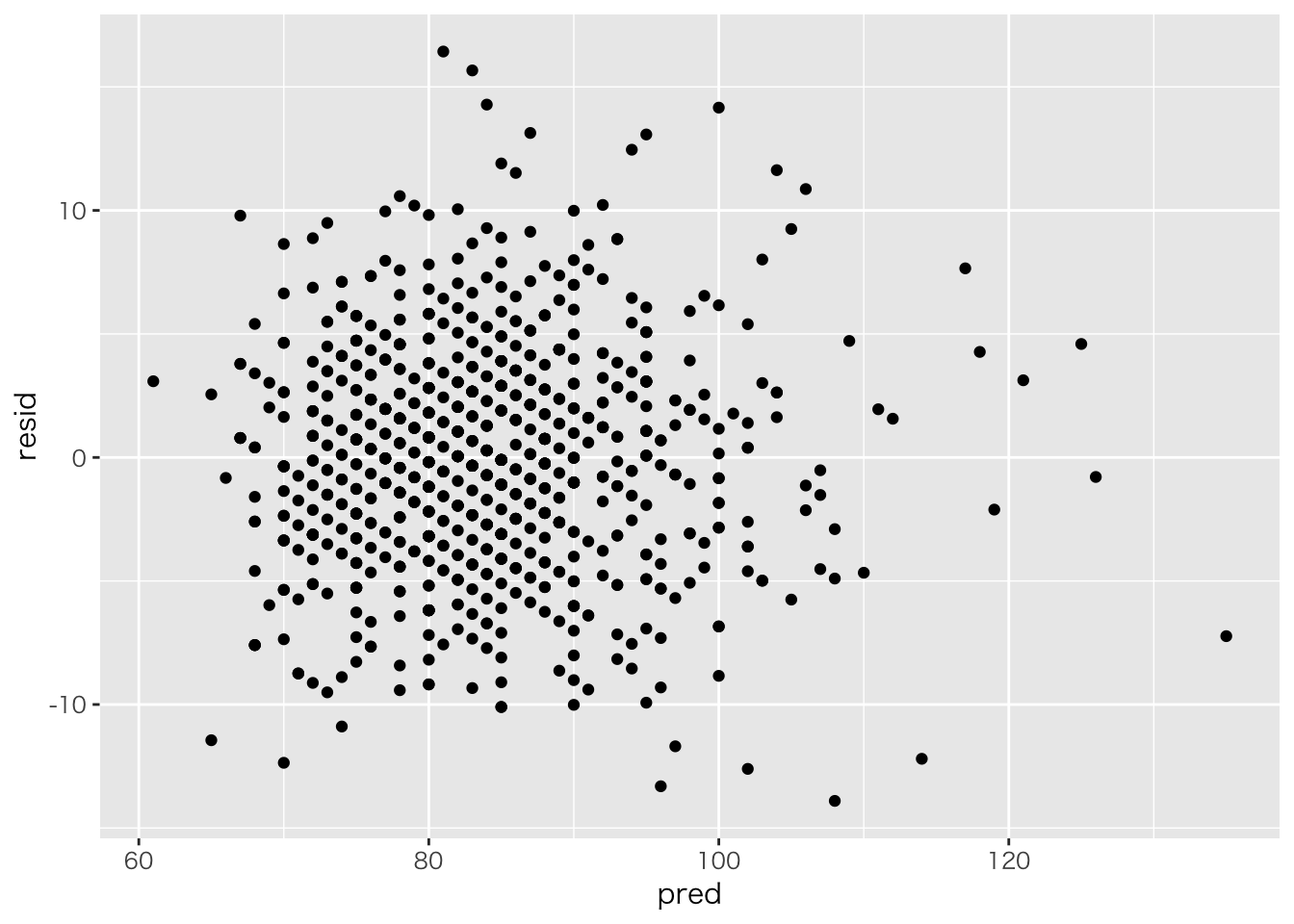 #### 予測値と残差の共分散
#### 予測値と残差の共分散
cov(result.lm$fitted.values,result.lm$residuals)## [1] -9.339304e-15data.frame(predicted=result.lm$fitted.values,resid=result.lm$residuals) %>%
ggplot(aes(x=predicted,y=resid))+geom_point()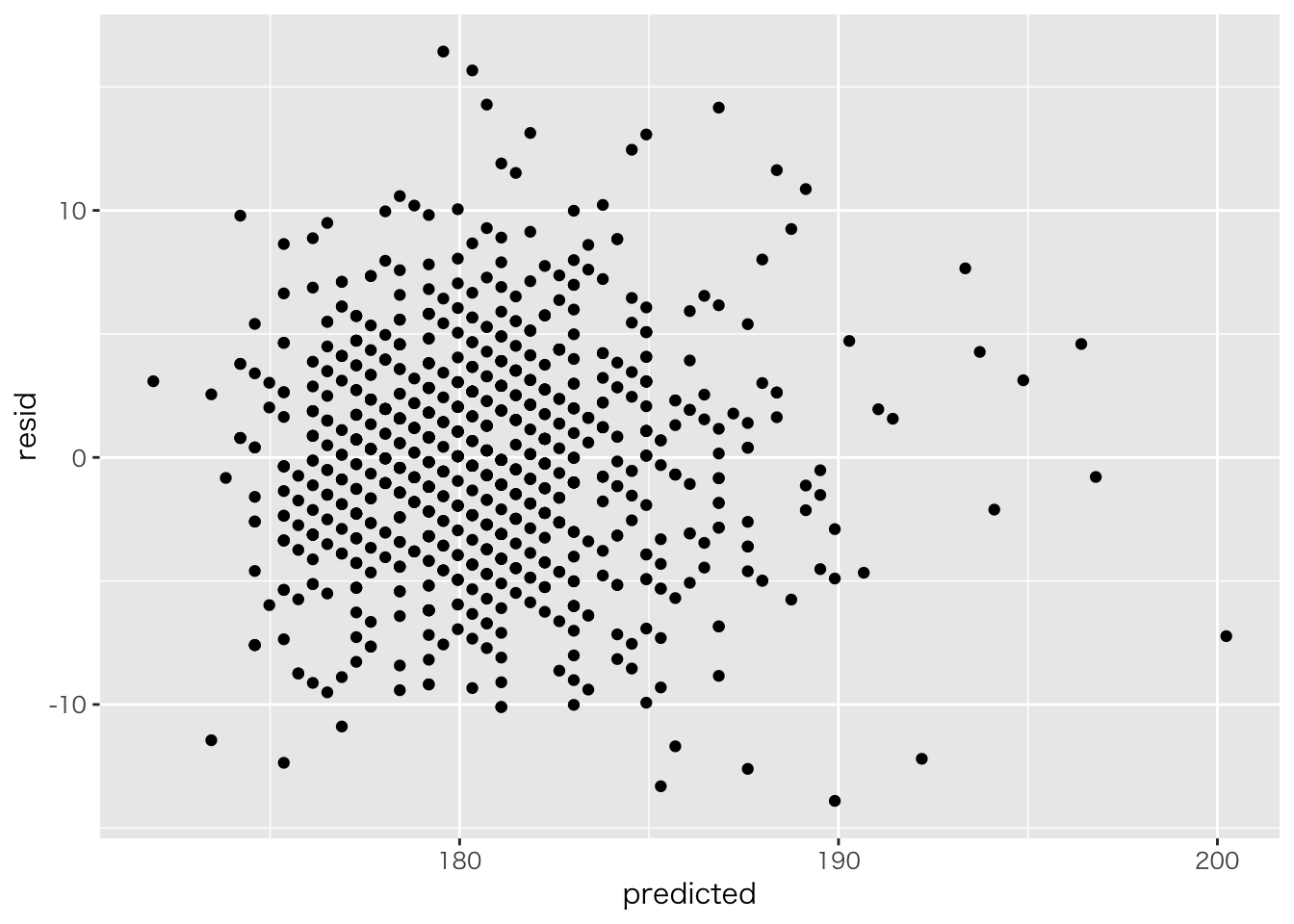
予測値と残差の共分散
var(baseball_dat$height)## [1] 31.20388var(result.lm$fitted.values)+var(result.lm$residuals)## [1] 31.20388予測値と被説明変数の共分散
cov(result.lm$fitted.values,baseball_dat$height)## [1] 12.12676var(result.lm$fitted.values)## [1] 12.12676課題3
- 野球データセットのうち,巨人群のデータだけ抽出して
- 体重を身長で予測する回帰分析を行い
- 回帰係数を報告しなさい。
##
## Call:
## lm(formula = weight ~ height, data = .)
##
## Residuals:
## Min 1Q Median 3Q Max
## -15.5839 -3.9816 -0.7728 3.0377 20.6872
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -125.9148 21.4783 -5.862 7.81e-08 ***
## height 1.1622 0.1186 9.802 9.17e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 6.688 on 88 degrees of freedom
## Multiple R-squared: 0.5219, Adjusted R-squared: 0.5165
## F-statistic: 96.08 on 1 and 88 DF, p-value: 9.174e-16線形モデル2;ベイズ推定
result.brms1 <- brm(height~weight,data=baseball_dat)## Compiling the C++ model## Start sampling##
## SAMPLING FOR MODEL 'f9a1272a26677f95a8c881fb5630f84c' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 0.000105 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 1.05 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 0.352193 seconds (Warm-up)
## Chain 1: 0.223614 seconds (Sampling)
## Chain 1: 0.575807 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL 'f9a1272a26677f95a8c881fb5630f84c' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 5.4e-05 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.54 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 0.432706 seconds (Warm-up)
## Chain 2: 0.302807 seconds (Sampling)
## Chain 2: 0.735513 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL 'f9a1272a26677f95a8c881fb5630f84c' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 3.9e-05 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0.39 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 3: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 3: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 3: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 3: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 3: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 3: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 3: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 3: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 3: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 0.291505 seconds (Warm-up)
## Chain 3: 0.215984 seconds (Sampling)
## Chain 3: 0.507489 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL 'f9a1272a26677f95a8c881fb5630f84c' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 4.1e-05 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0.41 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 4: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 4: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 4: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 4: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 4: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 4: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 4: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 4: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 4: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 0.398796 seconds (Warm-up)
## Chain 4: 0.275083 seconds (Sampling)
## Chain 4: 0.673879 seconds (Total)
## Chain 4:result.brms1## Family: gaussian
## Links: mu = identity; sigma = identity
## Formula: height ~ weight
## Data: baseball_dat (Number of observations: 918)
## Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup samples = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
## Intercept 148.58 1.33 146.00 151.22 3745 1.00
## weight 0.38 0.02 0.35 0.41 3666 1.00
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
## sigma 4.37 0.11 4.17 4.59 3274 1.00
##
## Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample
## is a crude measure of effective sample size, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).推定後のチェック
plot(result.brms1)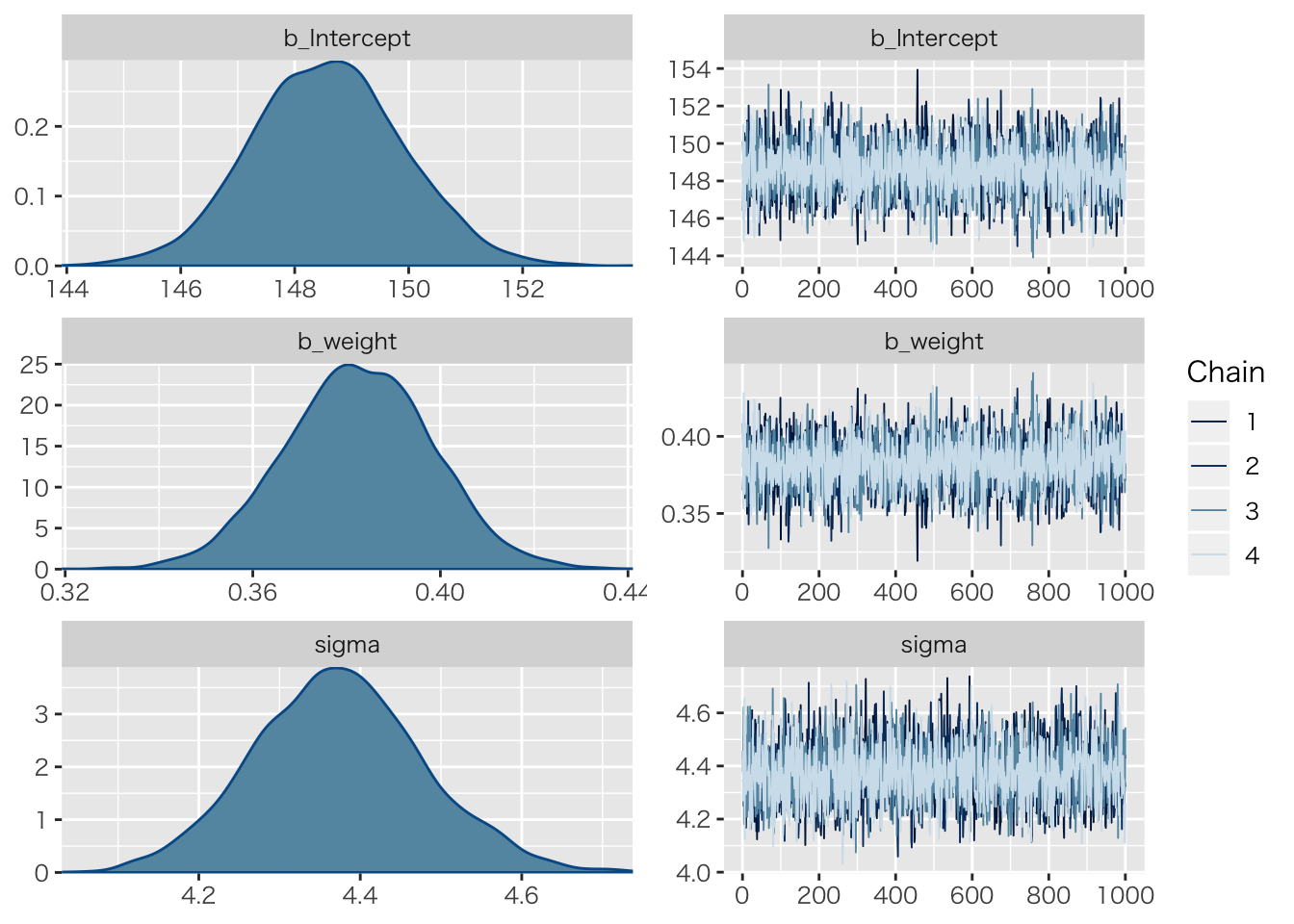
MCMCサンプルを見てみる
rstan::extract(result.brms1$fit) %>% data.frame %>% head()| b_Intercept | b | sigma | lp__ |
|---|---|---|---|
| 150.3875 | 0.3638039 | 4.349472 | -2662.044 |
| 146.7242 | 0.4032812 | 4.469978 | -2661.634 |
| 148.7295 | 0.3828856 | 4.300118 | -2660.987 |
| 148.7658 | 0.3797371 | 4.164917 | -2662.232 |
| 149.6226 | 0.3690926 | 4.254566 | -2661.234 |
| 147.0189 | 0.4007485 | 4.278681 | -2661.111 |
回帰直線
plot(marginal_effects(result.brms1))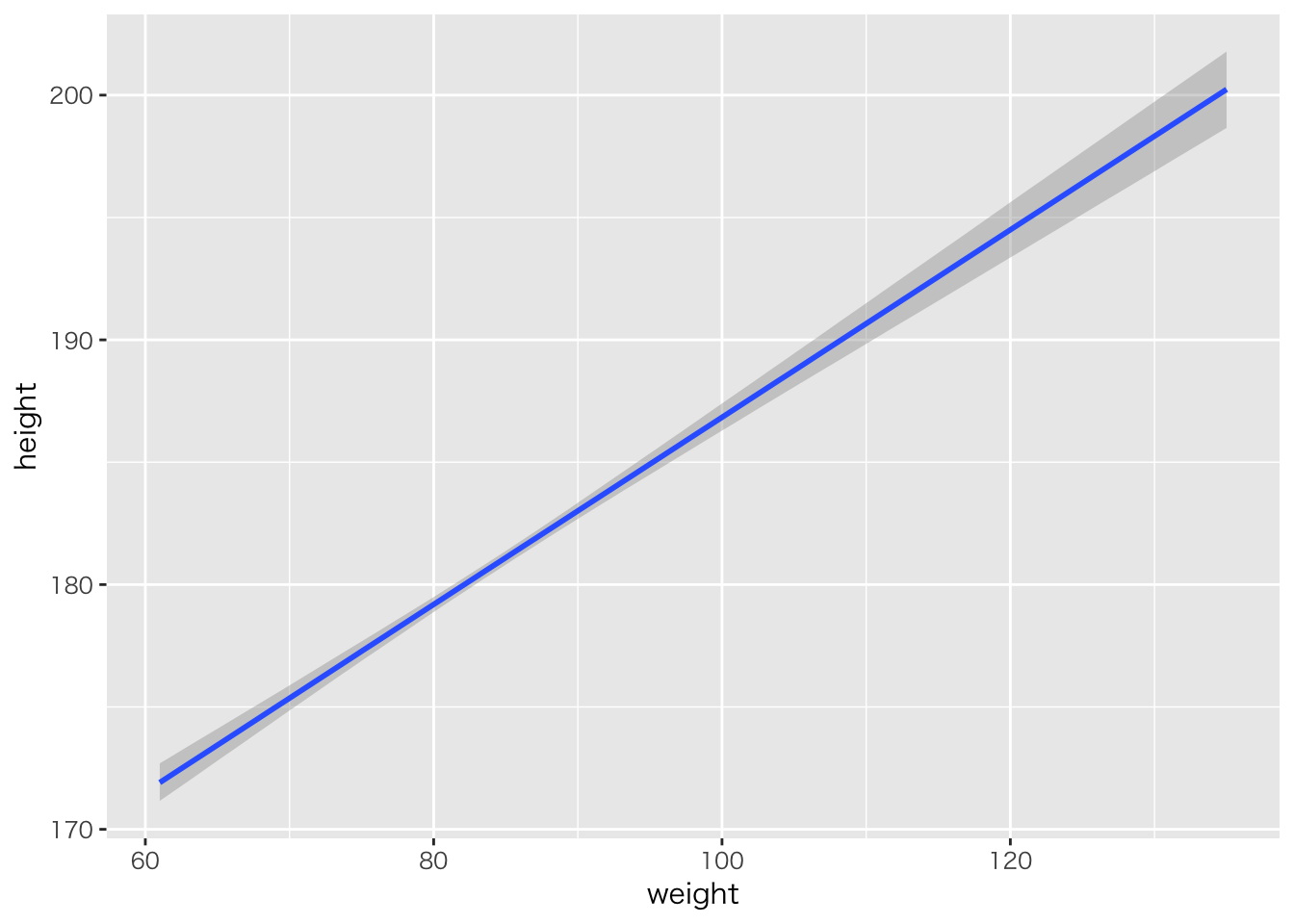
推定値
result.brms1 %>% fitted() %>% head()## Estimate Est.Error Q2.5 Q97.5
## [1,] 175.7423 0.2522298 175.2598 176.2452
## [2,] 181.0990 0.1435555 180.8152 181.3749
## [3,] 180.3337 0.1442661 180.0464 180.6146
## [4,] 179.5685 0.1516484 179.2721 179.8631
## [5,] 182.2468 0.1549641 181.9451 182.5472
## [6,] 183.0120 0.1695858 182.6829 183.3426事後予測分布
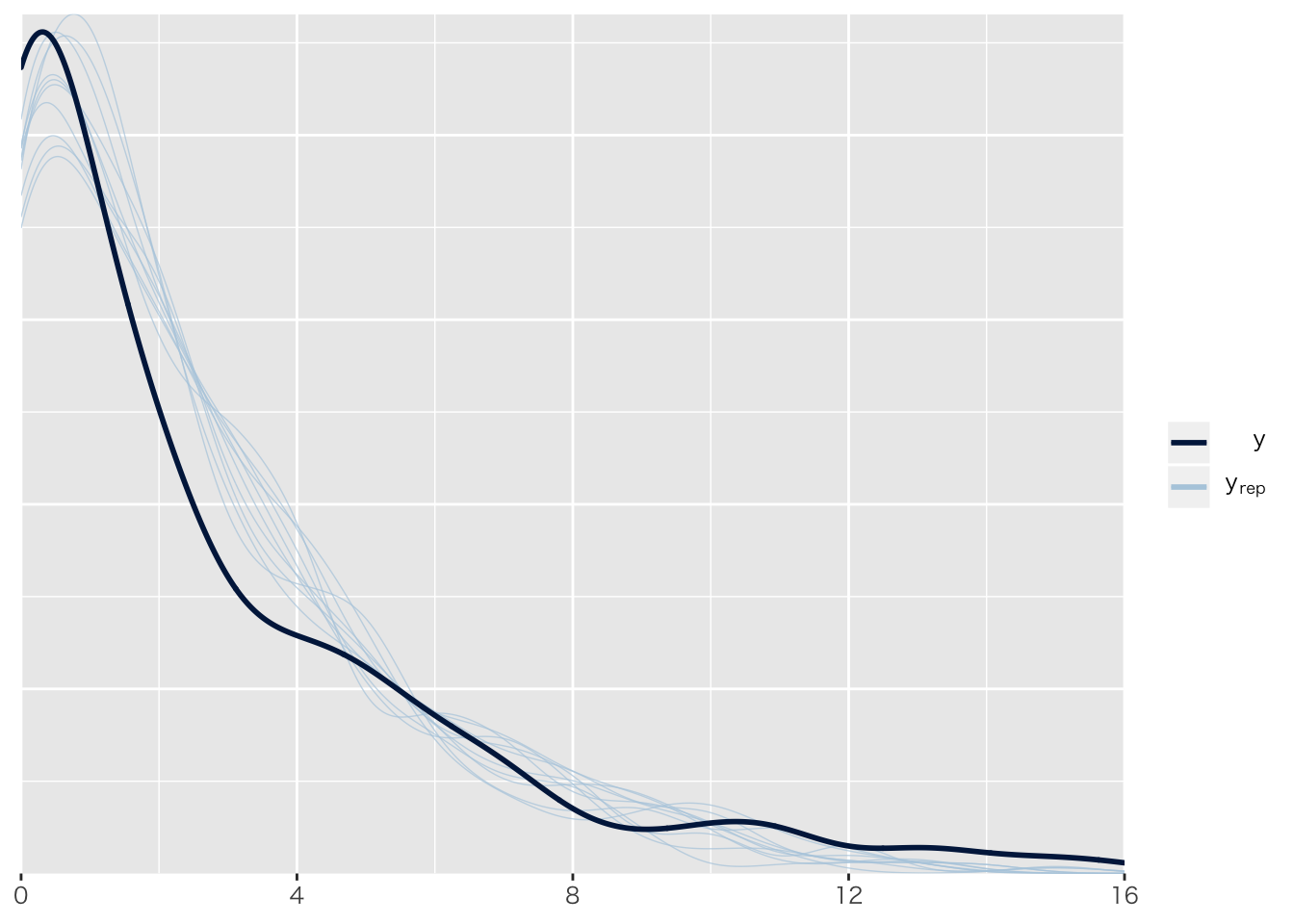
pp_check(result.brms1)## Using 10 posterior samples for ppc type 'dens_overlay' by default.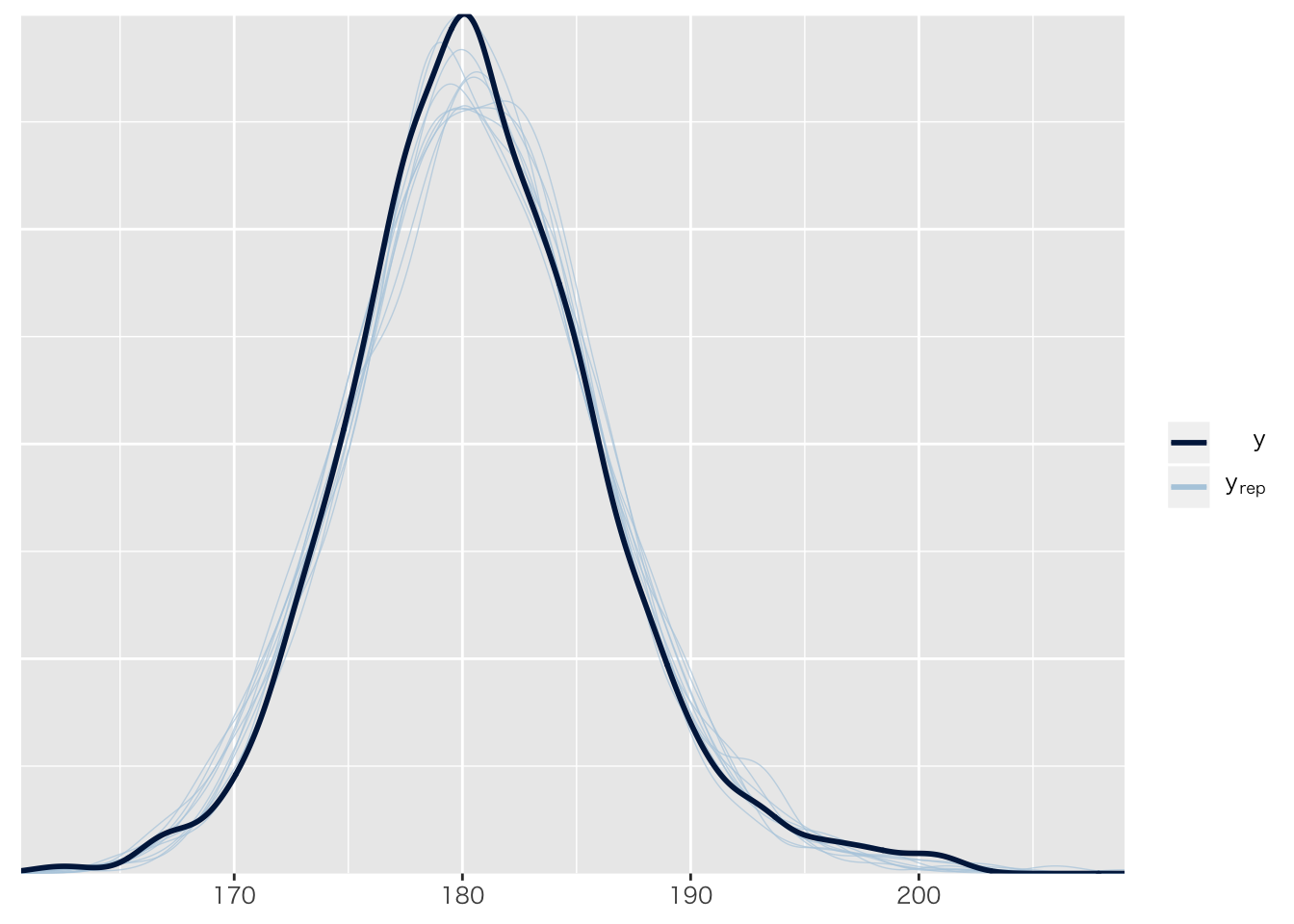
事後予測分布のサンプル
predict(result.brms1) %>% head(10)## Estimate Est.Error Q2.5 Q97.5
## [1,] 175.7529 4.395169 167.0688 184.1624
## [2,] 181.0027 4.393014 172.6260 189.8314
## [3,] 180.2229 4.392690 171.6723 188.6191
## [4,] 179.5035 4.373217 171.0744 187.9064
## [5,] 182.2918 4.344601 173.9330 190.8557
## [6,] 183.0462 4.368775 174.2325 191.5648
## [7,] 185.2292 4.370180 176.6598 193.8927
## [8,] 181.1025 4.410562 172.3840 189.7241
## [9,] 183.8497 4.406048 175.3437 192.5573
## [10,] 183.1184 4.398847 174.3461 192.0448チェックしておこう
- MCMCサンプルの数,チェイン数などMCMC推定オプション
- 事前分布の設定の仕方。
brm(height~weight,
data=baseball_dat,
iter = 2000,
warmup = 1000,
seed = 1234,
chain = 4)
?brms::set_prior課題4
- 課題3と同じ分析をベイズ推定しなさい
##
## SAMPLING FOR MODEL '9192c5613846522e9b8560b085cd9956' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 7.2e-05 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.72 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 0.067413 seconds (Warm-up)
## Chain 1: 0.057435 seconds (Sampling)
## Chain 1: 0.124848 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL '9192c5613846522e9b8560b085cd9956' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 1.6e-05 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.16 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 0.070683 seconds (Warm-up)
## Chain 2: 0.065265 seconds (Sampling)
## Chain 2: 0.135948 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL '9192c5613846522e9b8560b085cd9956' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 1.5e-05 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0.15 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 3: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 3: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 3: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 3: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 3: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 3: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 3: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 3: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 3: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 0.074083 seconds (Warm-up)
## Chain 3: 0.055762 seconds (Sampling)
## Chain 3: 0.129845 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL '9192c5613846522e9b8560b085cd9956' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 1.3e-05 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0.13 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 4: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 4: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 4: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 4: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 4: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 4: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 4: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 4: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 4: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 0.061684 seconds (Warm-up)
## Chain 4: 0.05435 seconds (Sampling)
## Chain 4: 0.116034 seconds (Total)
## Chain 4:## Family: gaussian
## Links: mu = identity; sigma = identity
## Formula: weight ~ height
## Data: . (Number of observations: 90)
## Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup samples = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
## Intercept -126.33 21.77 -168.32 -84.18 4307 1.00
## height 1.16 0.12 0.93 1.40 4274 1.00
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
## sigma 6.77 0.54 5.84 7.93 3215 1.00
##
## Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample
## is a crude measure of effective sample size, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).線形モデル3;一般線形モデル
利き腕の違いを考慮したいとする。
read_csv('baseball2019.csv') %>%
dplyr::select(height,weight,throw.by) %>%
na.omit() %>%
dplyr::mutate(throw.by=as.factor(throw.by)) -> baseball_dat3## Parsed with column specification:
## cols(
## .default = col_double(),
## Name = col_character(),
## team = col_character(),
## position = col_character(),
## bloodType = col_character(),
## throw.by = col_character(),
## batting.by = col_character(),
## birth.place = col_character(),
## birth.day = col_date(format = ""),
## 背番号 = col_character()
## )## See spec(...) for full column specifications.baseball_dat3 %>%
ggplot(aes(x=throw.by,y=height))+geom_point()+stat_summary(fun.y=mean,geom='line',group=1)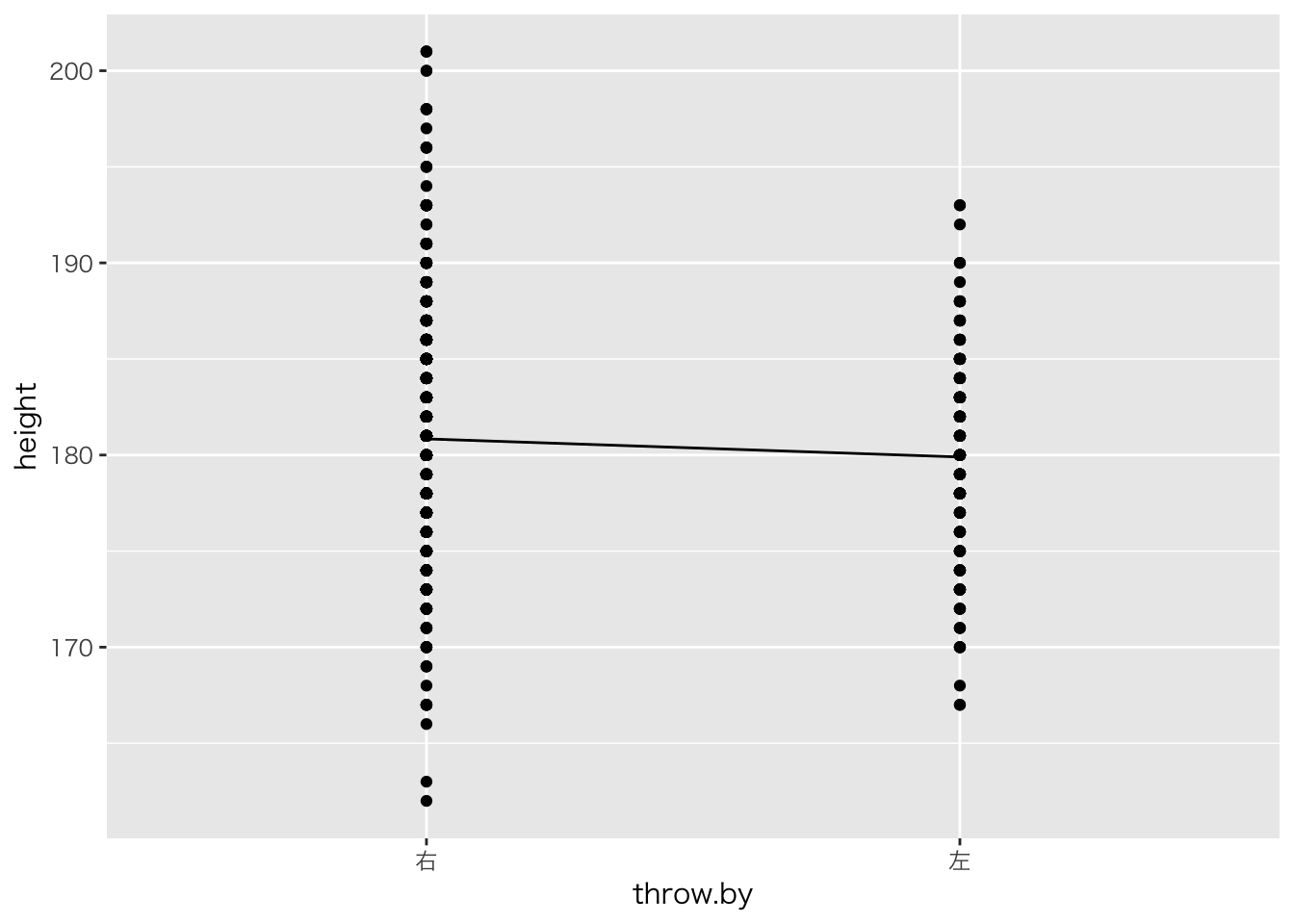
回帰分析でOK。t検定の別解でもある。
result.lm4 <- lm(height~throw.by,baseball_dat3)
summary(result.lm4)##
## Call:
## lm(formula = height ~ throw.by, data = baseball_dat3)
##
## Residuals:
## Min 1Q Median 3Q Max
## -18.8455 -3.8455 -0.8455 3.1545 20.1545
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 180.8455 0.2035 888.626 <2e-16 ***
## throw.by左 -0.9473 0.4771 -1.985 0.0474 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 5.577 on 916 degrees of freedom
## Multiple R-squared: 0.004285, Adjusted R-squared: 0.003198
## F-statistic: 3.942 on 1 and 916 DF, p-value: 0.0474ちなみに検定では
t.test(height~throw.by,data=baseball_dat3)##
## Welch Two Sample t-test
##
## data: height by throw.by
## t = 2.0617, df = 255.96, p-value = 0.04025
## alternative hypothesis: true difference in means is not equal to 0
## 95 percent confidence interval:
## 0.04245972 1.85221166
## sample estimates:
## mean in group 右 mean in group 左
## 180.8455 179.8982ベイズ推定では
こっちのほうがわかりやすい。
result.brms2 <- brm(height~throw.by,baseball_dat3)## Compiling the C++ model## recompiling to avoid crashing R session## Start sampling##
## SAMPLING FOR MODEL 'f9a1272a26677f95a8c881fb5630f84c' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 8.1e-05 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.81 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 0.355082 seconds (Warm-up)
## Chain 1: 0.164915 seconds (Sampling)
## Chain 1: 0.519997 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL 'f9a1272a26677f95a8c881fb5630f84c' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 4.4e-05 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.44 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 0.436207 seconds (Warm-up)
## Chain 2: 0.22492 seconds (Sampling)
## Chain 2: 0.661127 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL 'f9a1272a26677f95a8c881fb5630f84c' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 4.4e-05 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0.44 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 3: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 3: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 3: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 3: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 3: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 3: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 3: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 3: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 3: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 0.432731 seconds (Warm-up)
## Chain 3: 0.262282 seconds (Sampling)
## Chain 3: 0.695013 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL 'f9a1272a26677f95a8c881fb5630f84c' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 4.2e-05 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0.42 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 4: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 4: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 4: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 4: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 4: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 4: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 4: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 4: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 4: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 0.357155 seconds (Warm-up)
## Chain 4: 0.346893 seconds (Sampling)
## Chain 4: 0.704048 seconds (Total)
## Chain 4:result.brms2## Family: gaussian
## Links: mu = identity; sigma = identity
## Formula: height ~ throw.by
## Data: baseball_dat3 (Number of observations: 918)
## Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup samples = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
## Intercept 180.85 0.20 180.47 181.24 4603 1.00
## throw.by左 -0.96 0.47 -1.87 -0.02 4320 1.00
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
## sigma 5.58 0.13 5.33 5.85 3797 1.00
##
## Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample
## is a crude measure of effective sample size, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).plot(marginal_effects(result.brms2))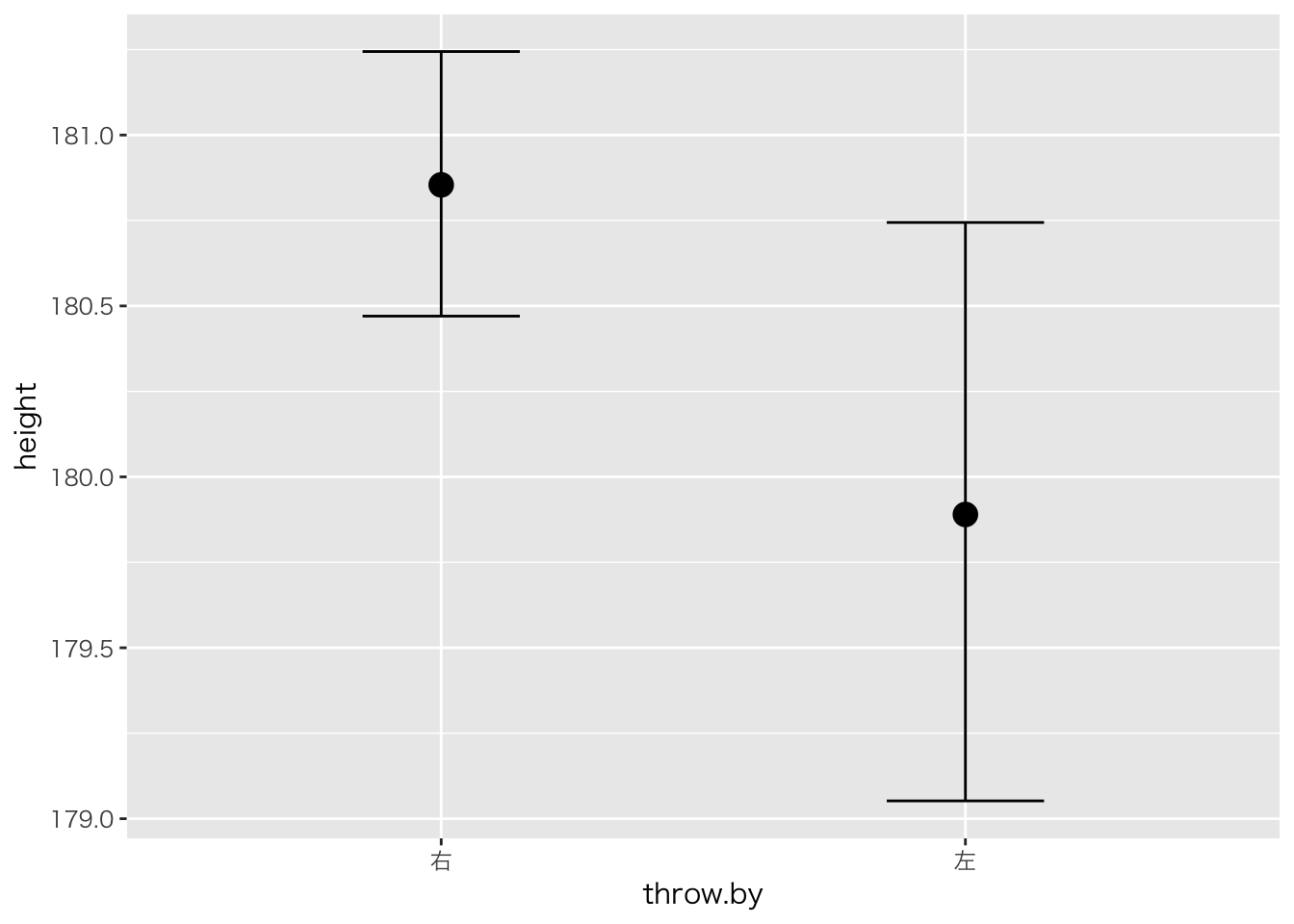
二要因の場合(間・間計画)
利き手は投げる時と打つときがあるので・・・
read_csv("baseball2019.csv") %>%
dplyr::select(height,weight,throw.by,batting.by) %>%
na.omit() %>%
dplyr::mutate(throw.by=as.factor(throw.by),
batting.by = as.factor(batting.by)) -> baseball_dat4## Parsed with column specification:
## cols(
## .default = col_double(),
## Name = col_character(),
## team = col_character(),
## position = col_character(),
## bloodType = col_character(),
## throw.by = col_character(),
## batting.by = col_character(),
## birth.place = col_character(),
## birth.day = col_date(format = ""),
## 背番号 = col_character()
## )## See spec(...) for full column specifications.result.brms4 <- brm(height ~ throw.by*batting.by,data=baseball_dat4)## Compiling the C++ model## recompiling to avoid crashing R session## Start sampling##
## SAMPLING FOR MODEL 'f9a1272a26677f95a8c881fb5630f84c' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 0.000104 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 1.04 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 2.9133 seconds (Warm-up)
## Chain 1: 2.43391 seconds (Sampling)
## Chain 1: 5.34721 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL 'f9a1272a26677f95a8c881fb5630f84c' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 6.2e-05 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.62 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 2.97995 seconds (Warm-up)
## Chain 2: 2.29487 seconds (Sampling)
## Chain 2: 5.27483 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL 'f9a1272a26677f95a8c881fb5630f84c' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 7.3e-05 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0.73 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 3: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 3: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 3: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 3: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 3: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 3: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 3: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 3: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 3: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 3.9114 seconds (Warm-up)
## Chain 3: 2.34817 seconds (Sampling)
## Chain 3: 6.25957 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL 'f9a1272a26677f95a8c881fb5630f84c' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 6e-05 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0.6 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 4: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 4: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 4: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 4: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 4: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 4: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 4: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 4: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 4: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 3.09466 seconds (Warm-up)
## Chain 4: 2.99537 seconds (Sampling)
## Chain 4: 6.09004 seconds (Total)
## Chain 4:result.brms4## Family: gaussian
## Links: mu = identity; sigma = identity
## Formula: height ~ throw.by * batting.by
## Data: baseball_dat4 (Number of observations: 918)
## Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup samples = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Eff.Sample
## Intercept 181.74 0.25 181.26 182.23 3146
## throw.by左 4.42 3.86 -3.06 12.08 1521
## batting.by左 -2.84 0.45 -3.73 -1.97 3076
## batting.by両 -1.61 1.19 -3.87 0.68 3510
## throw.by左:batting.by左 -3.54 3.89 -11.29 3.99 1531
## throw.by左:batting.by両 0.43 6.84 -12.15 14.28 1908
## Rhat
## Intercept 1.00
## throw.by左 1.00
## batting.by左 1.00
## batting.by両 1.00
## throw.by左:batting.by左 1.00
## throw.by左:batting.by両 1.00
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
## sigma 5.46 0.13 5.23 5.71 3611 1.00
##
## Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample
## is a crude measure of effective sample size, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).plot(marginal_effects(result.brms4,effects="throw.by"))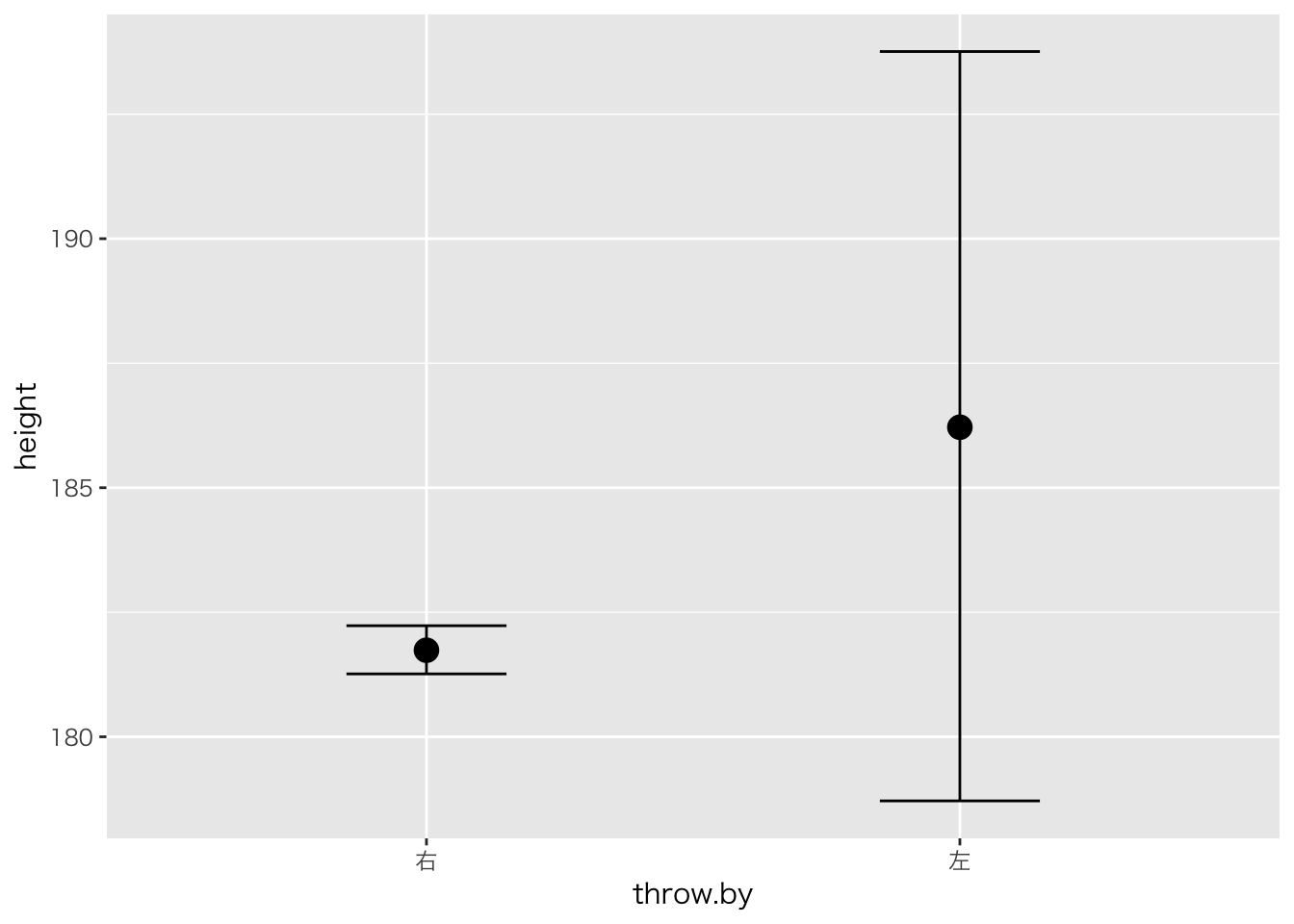
plot(marginal_effects(result.brms4,effects="batting.by"))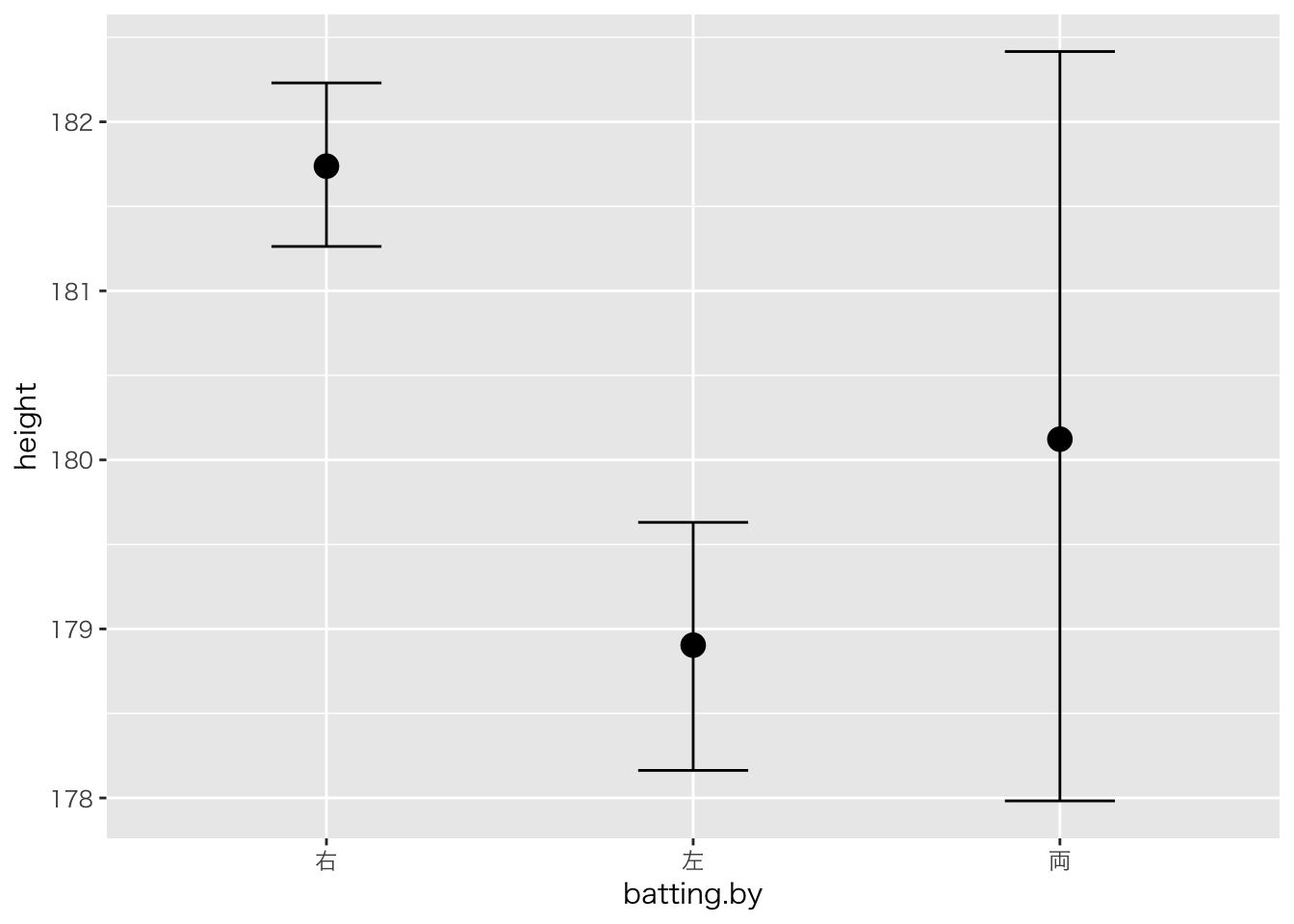
plot(marginal_effects(result.brms4,effects="throw.by:batting.by"))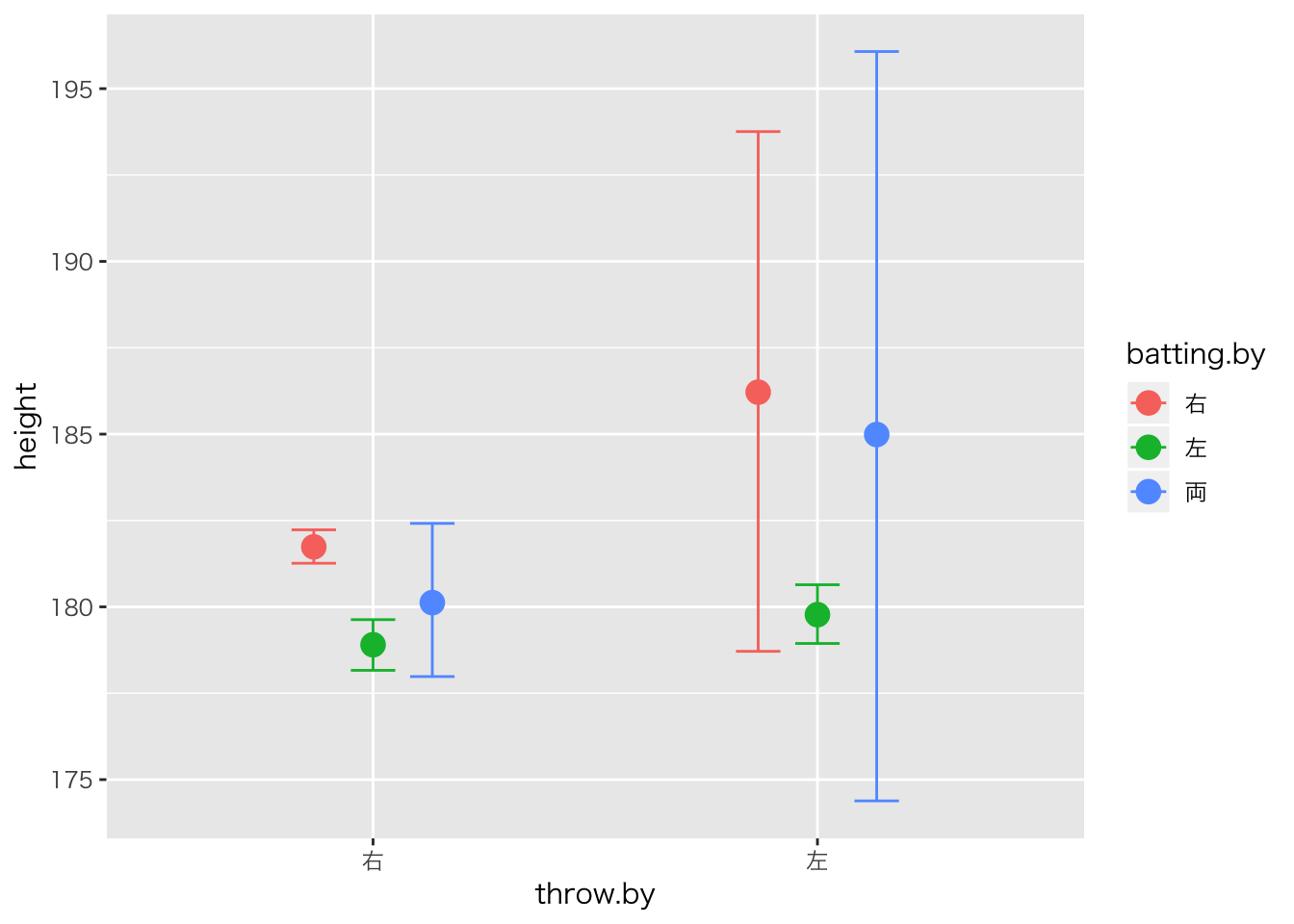
課題5
- 野球選手の体重について,利き腕の違いによる差があるかどうかを検証するため,
- t検定,最尤推定,ベイズ推定,それぞれの分析結果を出しなさい。
##
## Welch Two Sample t-test
##
## data: weight by throw.by
## t = 1.47, df = 266.26, p-value = 0.1427
## alternative hypothesis: true difference in means is not equal to 0
## 95 percent confidence interval:
## -0.3619578 2.4951295
## sample estimates:
## mean in group 右 mean in group 左
## 84.07856 83.01198##
## Call:
## lm(formula = weight ~ throw.by, data = baseball_dat3)
##
## Residuals:
## Min 1Q Median 3Q Max
## -23.079 -6.079 -1.012 4.921 50.921
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 84.0786 0.3319 253.320 <2e-16 ***
## throw.by左 -1.0666 0.7782 -1.371 0.171
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 9.096 on 916 degrees of freedom
## Multiple R-squared: 0.002047, Adjusted R-squared: 0.0009572
## F-statistic: 1.879 on 1 and 916 DF, p-value: 0.1708##
## SAMPLING FOR MODEL '121bc530f45cecede62dd01594d4df33' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 8.3e-05 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.83 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 0.355782 seconds (Warm-up)
## Chain 1: 0.302804 seconds (Sampling)
## Chain 1: 0.658586 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL '121bc530f45cecede62dd01594d4df33' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 7.4e-05 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.74 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 0.420004 seconds (Warm-up)
## Chain 2: 0.252997 seconds (Sampling)
## Chain 2: 0.673001 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL '121bc530f45cecede62dd01594d4df33' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 4.5e-05 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0.45 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 3: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 3: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 3: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 3: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 3: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 3: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 3: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 3: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 3: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 0.453687 seconds (Warm-up)
## Chain 3: 0.40502 seconds (Sampling)
## Chain 3: 0.858707 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL '121bc530f45cecede62dd01594d4df33' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 5.2e-05 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0.52 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 4: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 4: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 4: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 4: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 4: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 4: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 4: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 4: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 4: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 0.380931 seconds (Warm-up)
## Chain 4: 0.712407 seconds (Sampling)
## Chain 4: 1.09334 seconds (Total)
## Chain 4:## Family: gaussian
## Links: mu = identity; sigma = identity
## Formula: weight ~ throw.by
## Data: baseball_dat3 (Number of observations: 918)
## Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup samples = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
## Intercept 84.08 0.34 83.41 84.73 3805 1.00
## throw.by左 -1.08 0.77 -2.60 0.41 3518 1.00
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
## sigma 9.10 0.21 8.69 9.53 3955 1.00
##
## Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample
## is a crude measure of effective sample size, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).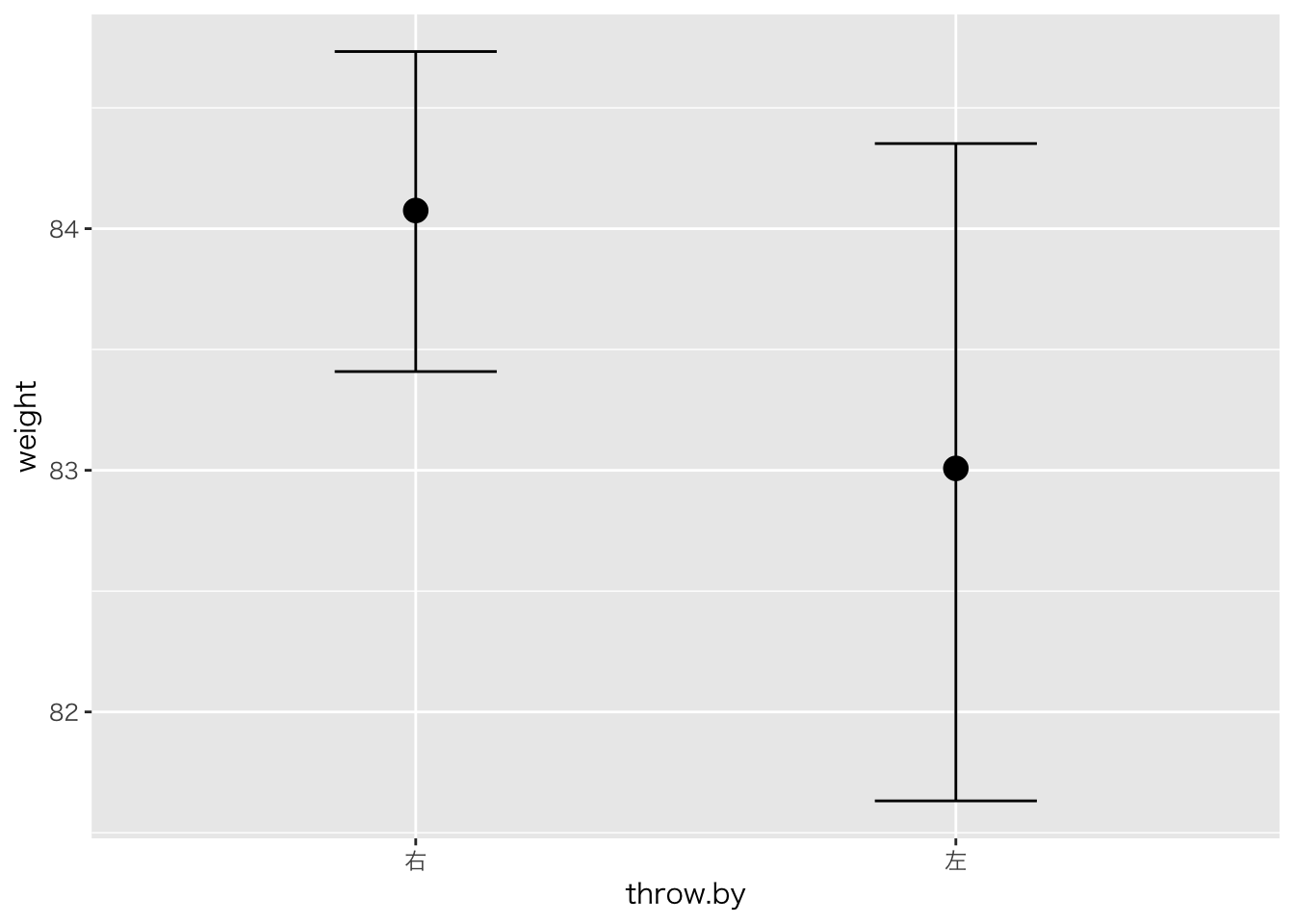
線形モデル4;一般化線形モデル
対数正規分布
年俸のデータは正規分布しない。
read_csv('baseball2019.csv') %>%
# 野手のデータ
dplyr::filter(position!="投手") %>%
ggplot(aes(x=salary))+geom_histogram(binwidth = 1000)## Parsed with column specification:
## cols(
## .default = col_double(),
## Name = col_character(),
## team = col_character(),
## position = col_character(),
## bloodType = col_character(),
## throw.by = col_character(),
## batting.by = col_character(),
## birth.place = col_character(),
## birth.day = col_date(format = ""),
## 背番号 = col_character()
## )## See spec(...) for full column specifications.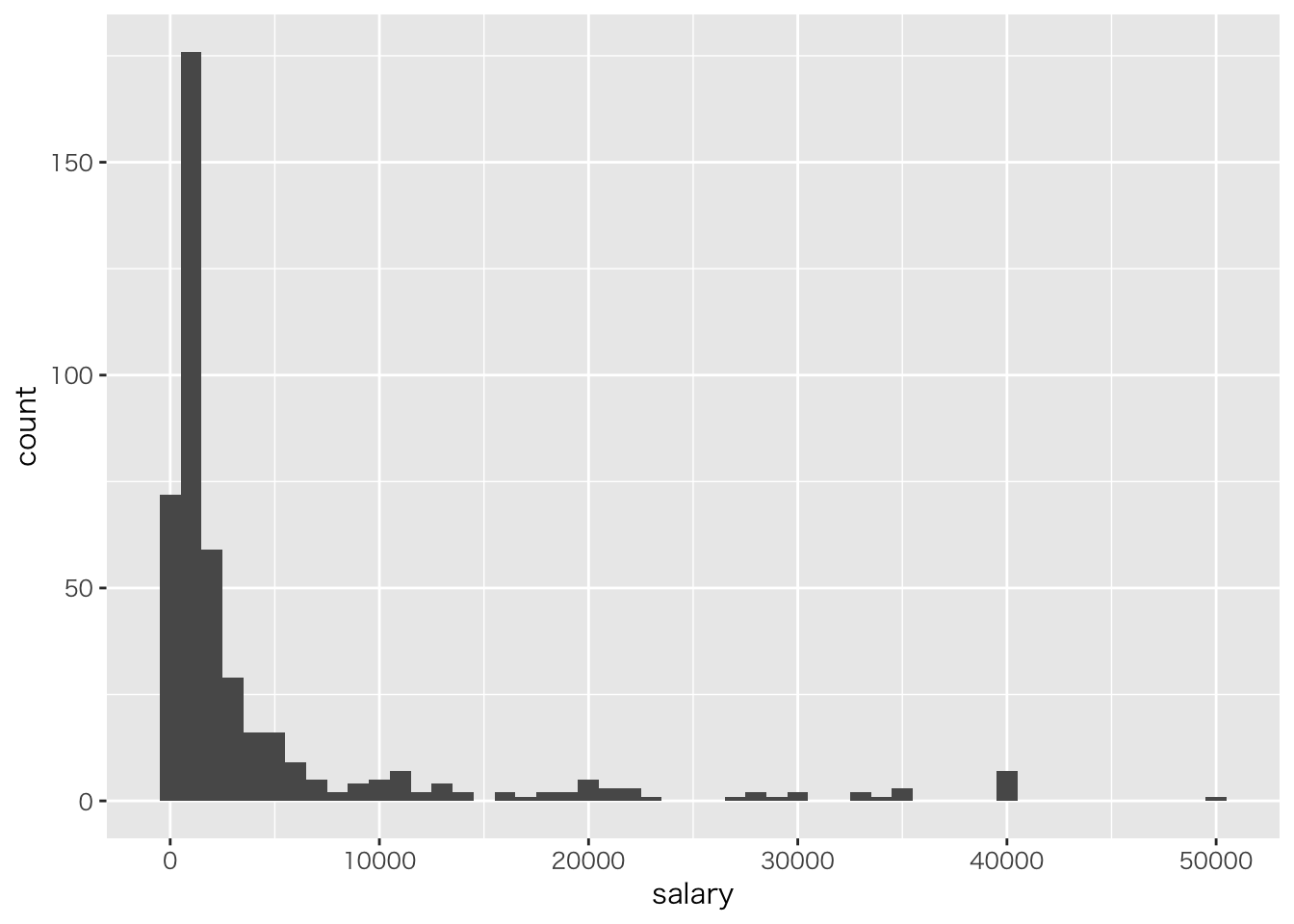
こういうデータに回帰線を当てるのはよくない。
read_csv('baseball2019.csv') %>%
# 野手のデータ
dplyr::filter(position!="投手") %>%
ggplot(aes(x=安打,y=salary))+geom_point()+geom_smooth(method='lm',se=F)## Parsed with column specification:
## cols(
## .default = col_double(),
## Name = col_character(),
## team = col_character(),
## position = col_character(),
## bloodType = col_character(),
## throw.by = col_character(),
## batting.by = col_character(),
## birth.place = col_character(),
## birth.day = col_date(format = ""),
## 背番号 = col_character()
## )## See spec(...) for full column specifications.## Warning: Removed 125 rows containing non-finite values (stat_smooth).## Warning: Removed 125 rows containing missing values (geom_point).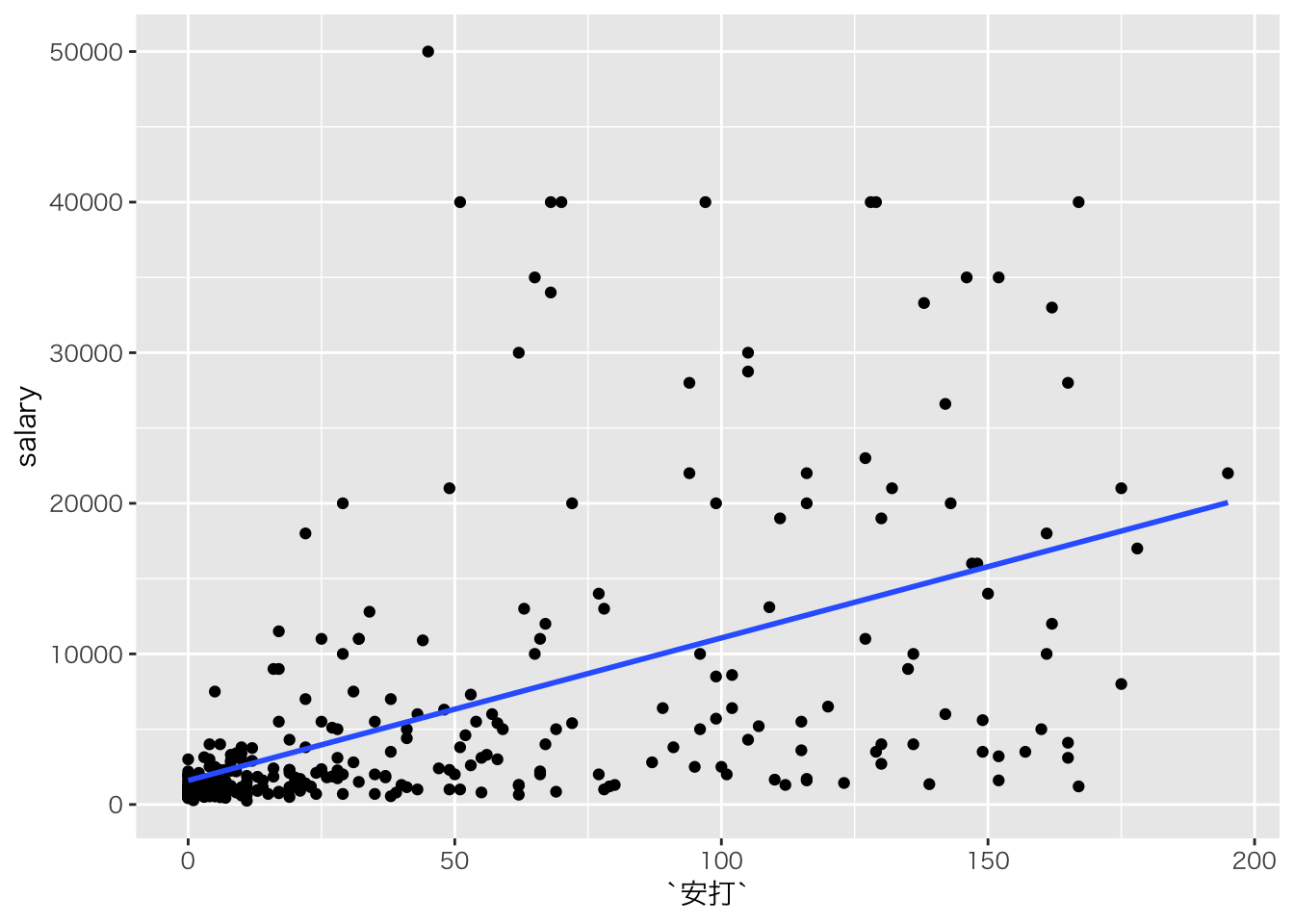
分布にあった分析をする。この場合は対数正規分布が良い。
ggplot(data.frame(x=c(0, 10)), aes(x=x)) + stat_function(fun = dlnorm)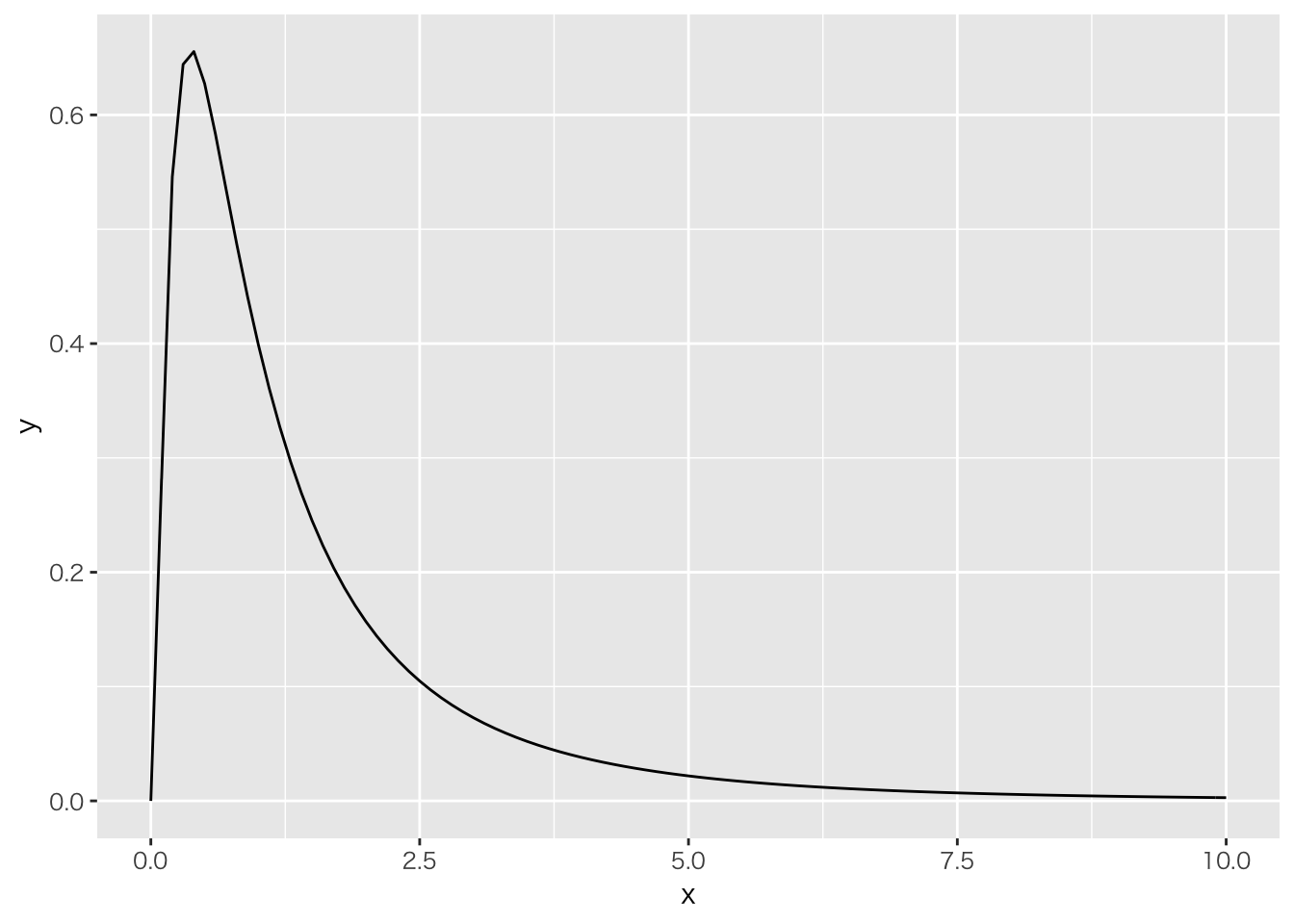
分析のコード
read_csv('baseball2019.csv') %>%
# 野手のデータ
dplyr::filter(position!="投手") %>%
# 年収の単位を少し落とす(見にくいので)
dplyr::mutate(salary=salary/1000) %>%
brm(salary~安打,data=.,family="lognormal") -> result.brms6## Parsed with column specification:
## cols(
## .default = col_double(),
## Name = col_character(),
## team = col_character(),
## position = col_character(),
## bloodType = col_character(),
## throw.by = col_character(),
## batting.by = col_character(),
## birth.place = col_character(),
## birth.day = col_date(format = ""),
## 背番号 = col_character()
## )## See spec(...) for full column specifications.## Warning: Rows containing NAs were excluded from the model.## Compiling the C++ model## Start sampling##
## SAMPLING FOR MODEL 'c174d626c8a5eb443f88f4a10a6d919e' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 6.1e-05 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.61 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 0.436417 seconds (Warm-up)
## Chain 1: 0.195528 seconds (Sampling)
## Chain 1: 0.631945 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL 'c174d626c8a5eb443f88f4a10a6d919e' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 3e-05 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.3 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 0.358331 seconds (Warm-up)
## Chain 2: 0.205708 seconds (Sampling)
## Chain 2: 0.564039 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL 'c174d626c8a5eb443f88f4a10a6d919e' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 6e-05 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0.6 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 3: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 3: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 3: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 3: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 3: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 3: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 3: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 3: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 3: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 0.446142 seconds (Warm-up)
## Chain 3: 0.212787 seconds (Sampling)
## Chain 3: 0.658929 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL 'c174d626c8a5eb443f88f4a10a6d919e' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 6e-05 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0.6 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 4: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 4: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 4: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 4: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 4: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 4: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 4: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 4: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 4: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 0.434915 seconds (Warm-up)
## Chain 4: 0.212289 seconds (Sampling)
## Chain 4: 0.647204 seconds (Total)
## Chain 4:pp_check(result.brms6)## Using 10 posterior samples for ppc type 'dens_overlay' by default.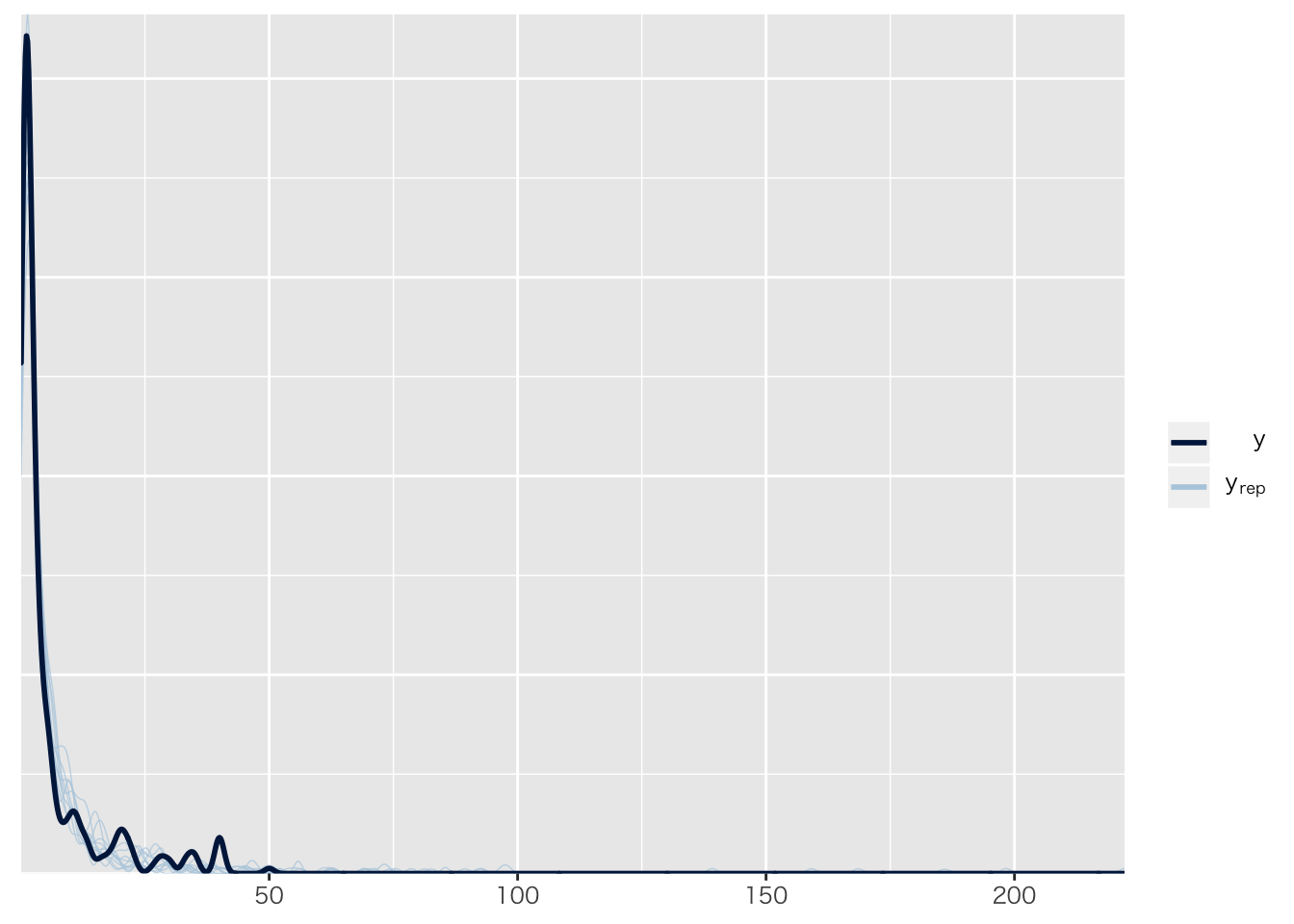 正規分布のままだとおかしいのは目に見えてわかる。
正規分布のままだとおかしいのは目に見えてわかる。
read_csv('baseball2019.csv') %>%
# 野手のデータ
dplyr::filter(position!="投手") %>%
# 年収の単位を少し落とす(見にくいので)
dplyr::mutate(salary=salary/1000) %>%
brm(salary~安打,data=.) %>%
pp_check()## Parsed with column specification:
## cols(
## .default = col_double(),
## Name = col_character(),
## team = col_character(),
## position = col_character(),
## bloodType = col_character(),
## throw.by = col_character(),
## batting.by = col_character(),
## birth.place = col_character(),
## birth.day = col_date(format = ""),
## 背番号 = col_character()
## )## See spec(...) for full column specifications.## Warning: Rows containing NAs were excluded from the model.## Compiling the C++ model## Start sampling##
## SAMPLING FOR MODEL 'da1510c7db6742ffc115392b5fbaec1b' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 6e-05 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.6 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 0.154702 seconds (Warm-up)
## Chain 1: 0.10641 seconds (Sampling)
## Chain 1: 0.261112 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL 'da1510c7db6742ffc115392b5fbaec1b' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 3e-05 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.3 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 0.14133 seconds (Warm-up)
## Chain 2: 0.137439 seconds (Sampling)
## Chain 2: 0.278769 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL 'da1510c7db6742ffc115392b5fbaec1b' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 2.4e-05 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0.24 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 3: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 3: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 3: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 3: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 3: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 3: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 3: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 3: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 3: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 0.124805 seconds (Warm-up)
## Chain 3: 0.122363 seconds (Sampling)
## Chain 3: 0.247168 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL 'da1510c7db6742ffc115392b5fbaec1b' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 2.3e-05 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0.23 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 4: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 4: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 4: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 4: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 4: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 4: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 4: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 4: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 4: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 0.144446 seconds (Warm-up)
## Chain 4: 0.112172 seconds (Sampling)
## Chain 4: 0.256618 seconds (Total)
## Chain 4:## Using 10 posterior samples for ppc type 'dens_overlay' by default.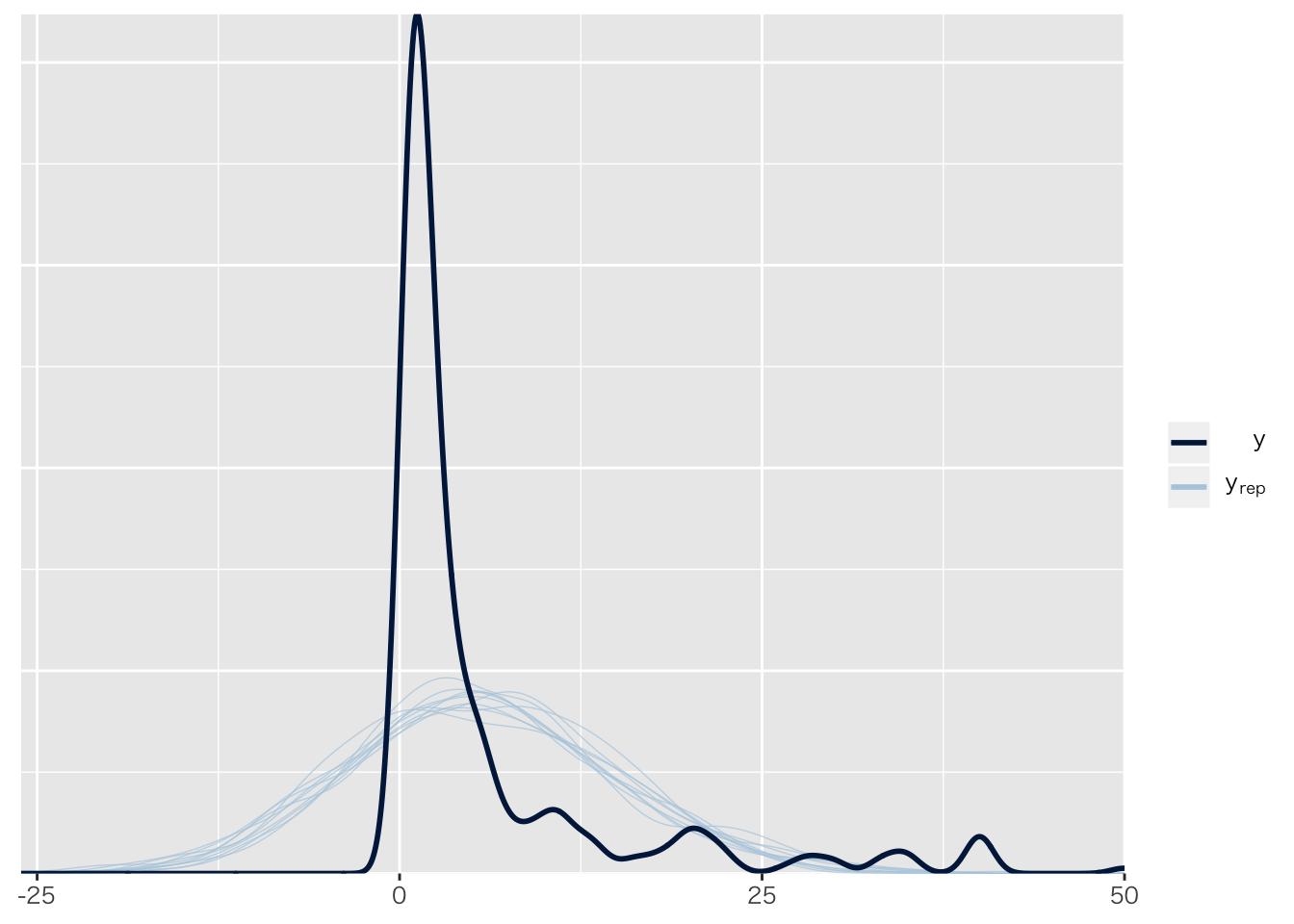
分析は分布に合わせないといけないことを肝に命じておいてほしい。
ポアソン分布
ggplot(data.frame(x=c(0:10)), aes(x)) +
stat_function(geom="point", n=11, fun=dpois, args=list(1),color="black") +
stat_function(geom="point", n=11, fun=dpois, args=list(3),color="red") +
stat_function(geom="point", n=11, fun=dpois, args=list(5),color="blue")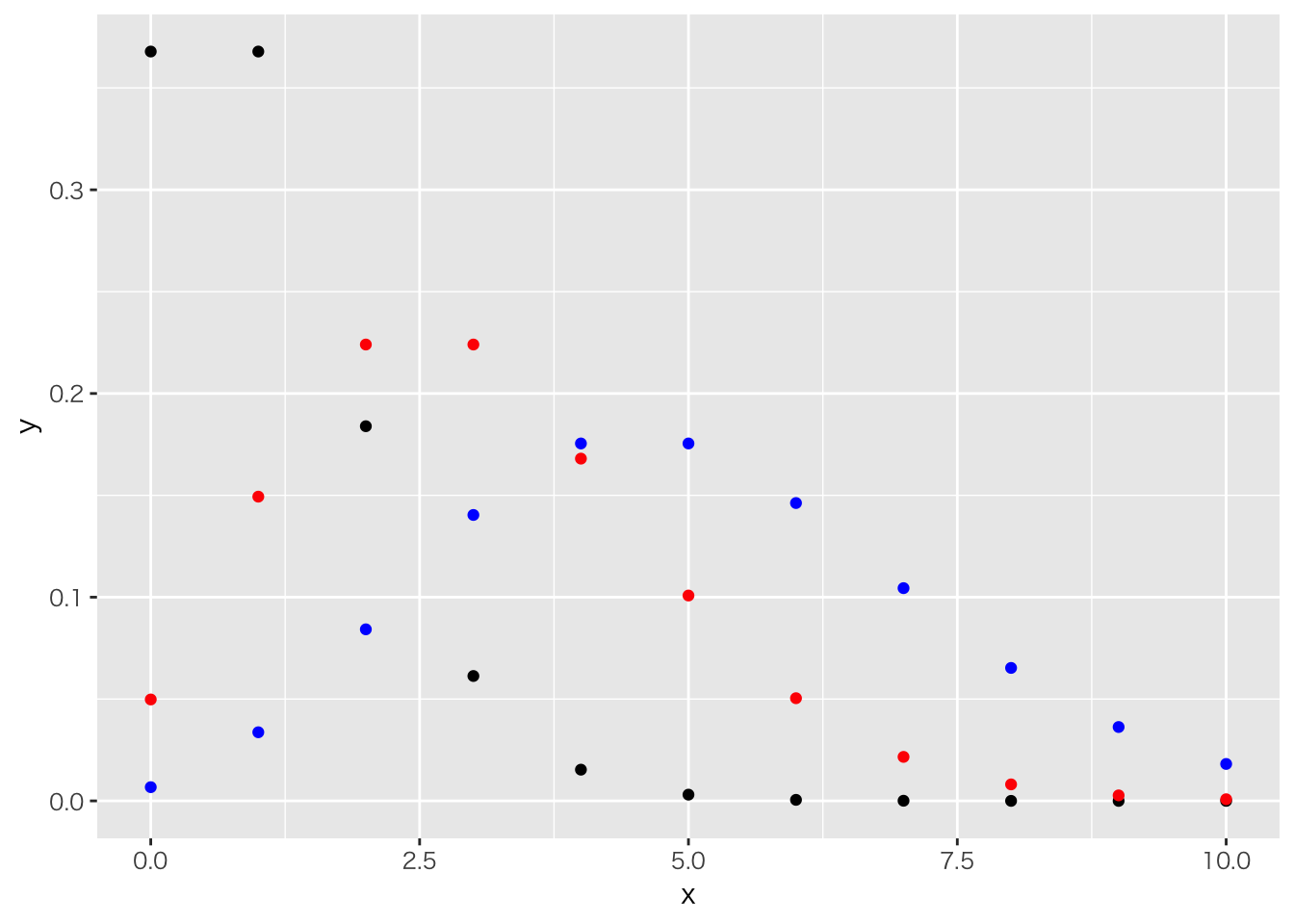
カウントデータに使う。
read_csv('baseball2019.csv') %>%
# 野手のデータ
dplyr::filter(position!="投手") %>%
dplyr::select(本塁打,weight) %>%
na.omit() -> baseball_dat6## Parsed with column specification:
## cols(
## .default = col_double(),
## Name = col_character(),
## team = col_character(),
## position = col_character(),
## bloodType = col_character(),
## throw.by = col_character(),
## batting.by = col_character(),
## birth.place = col_character(),
## birth.day = col_date(format = ""),
## 背番号 = col_character()
## )## See spec(...) for full column specifications.baseball_dat6 %>%
ggplot(aes(x=本塁打))+geom_histogram(binwidth=3)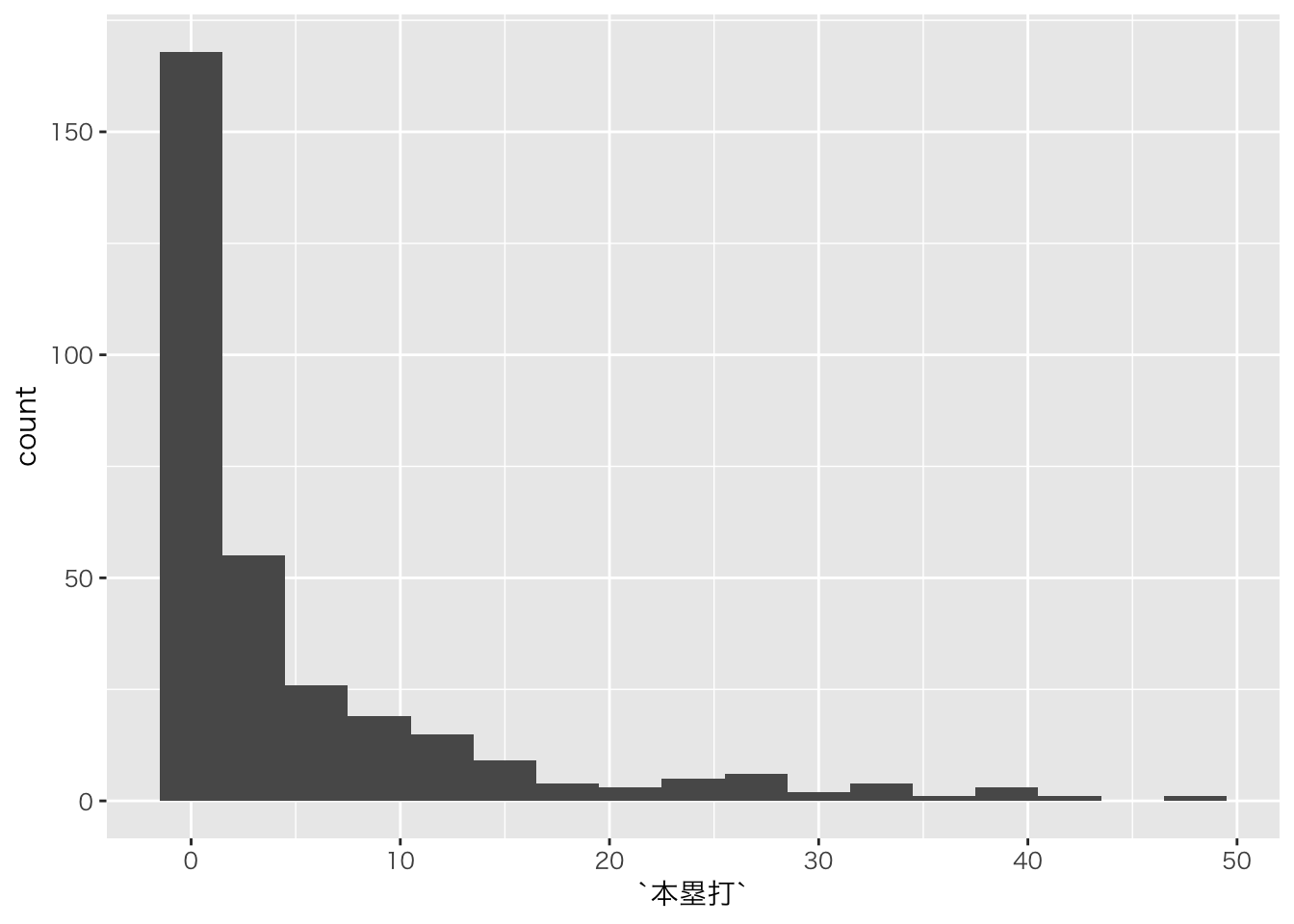
線形モデルの例。
result.brms7 <- brm(本塁打~weight,data=baseball_dat6,family="poisson")## Compiling the C++ model## Start sampling##
## SAMPLING FOR MODEL '84e3046c0af1517d6481f0c9f6abbda4' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 9.3e-05 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.93 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 0.520852 seconds (Warm-up)
## Chain 1: 0.348999 seconds (Sampling)
## Chain 1: 0.869851 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL '84e3046c0af1517d6481f0c9f6abbda4' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 6e-05 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.6 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 0.418863 seconds (Warm-up)
## Chain 2: 0.360305 seconds (Sampling)
## Chain 2: 0.779168 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL '84e3046c0af1517d6481f0c9f6abbda4' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 5.8e-05 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0.58 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 3: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 3: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 3: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 3: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 3: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 3: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 3: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 3: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 3: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 0.486497 seconds (Warm-up)
## Chain 3: 0.321302 seconds (Sampling)
## Chain 3: 0.807799 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL '84e3046c0af1517d6481f0c9f6abbda4' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 6.2e-05 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0.62 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 4: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 4: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 4: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 4: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 4: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 4: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 4: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 4: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 4: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 0.509766 seconds (Warm-up)
## Chain 4: 0.270811 seconds (Sampling)
## Chain 4: 0.780577 seconds (Total)
## Chain 4:result.brms7## Family: poisson
## Links: mu = log
## Formula: 本塁打 ~ weight
## Data: baseball_dat6 (Number of observations: 322)
## Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup samples = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
## Intercept -2.68 0.15 -2.98 -2.39 1964 1.00
## weight 0.05 0.00 0.05 0.05 2424 1.00
##
## Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample
## is a crude measure of effective sample size, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).pp_check(result.brms7)## Using 10 posterior samples for ppc type 'dens_overlay' by default.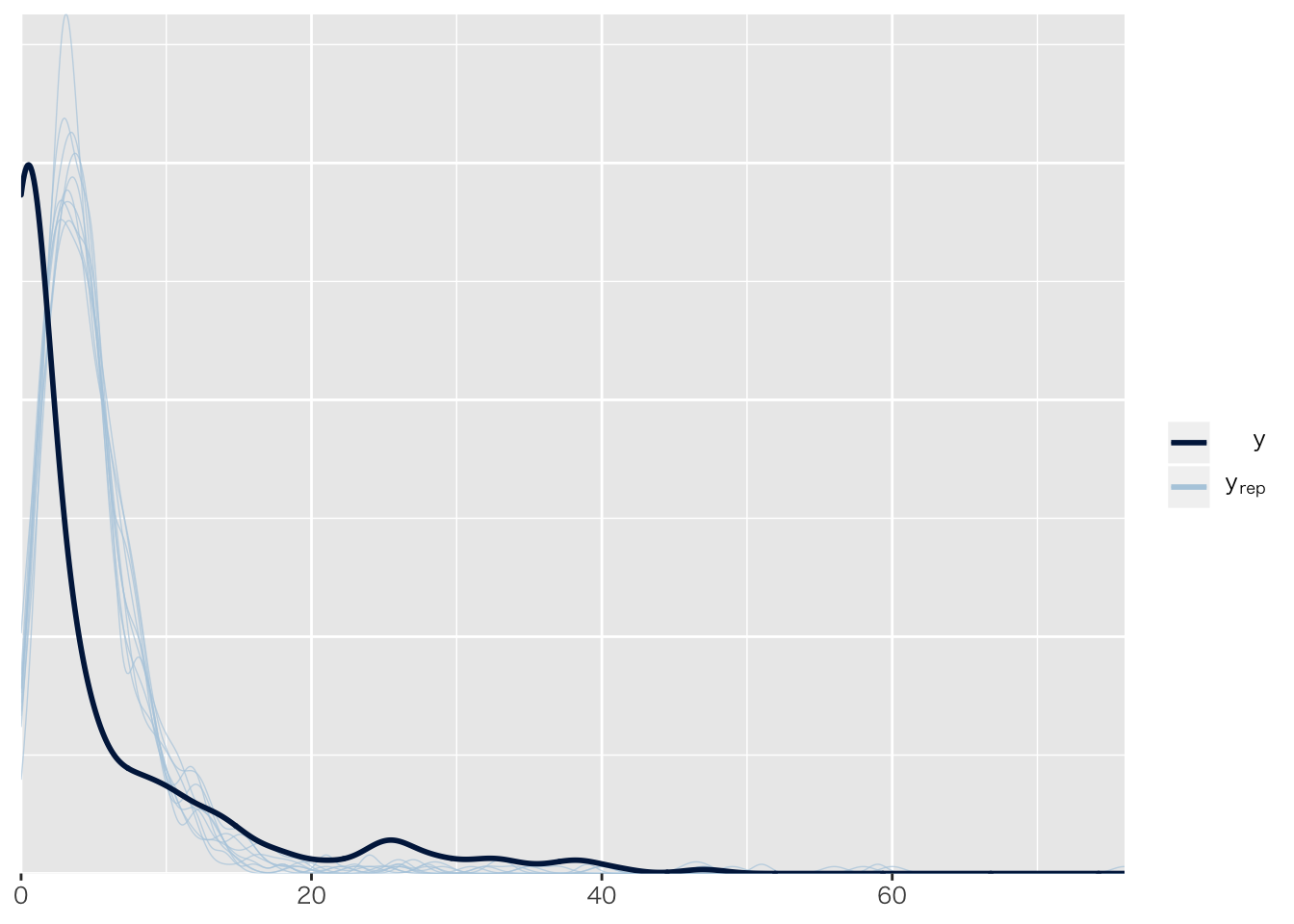
二項分布
ggplot(data.frame(x=c(0, 10)), aes(x=x)) +
stat_function(geom = "point",n=11,fun = dbinom,args=list(size=10,prob=0.5),color="black")+
stat_function(geom = "point",n=11,fun = dbinom,args=list(size=10,prob=0.3),color="red")+
stat_function(geom = "point",n=11,fun = dbinom,args=list(size=10,prob=0.7),color="blue")
割合のデータに使う。
read_csv('baseball2019.csv') %>%
# 野手のデータ
dplyr::filter(position!="投手") %>%
dplyr::select(打率,安打,打数,weight) %>%
# 日本語名を直しておく
dplyr::rename(BA=打率,HIT=安打,AB=打数) %>%
# 欠損値は除外
na.omit() %>%
# 打席に立っていない人のデータも除外
dplyr::filter(BA!=0) -> baseball_dat7## Parsed with column specification:
## cols(
## .default = col_double(),
## Name = col_character(),
## team = col_character(),
## position = col_character(),
## bloodType = col_character(),
## throw.by = col_character(),
## batting.by = col_character(),
## birth.place = col_character(),
## birth.day = col_date(format = ""),
## 背番号 = col_character()
## )## See spec(...) for full column specifications.baseball_dat7 %>%
ggplot(aes(x=BA))+geom_histogram(binwidth=0.01)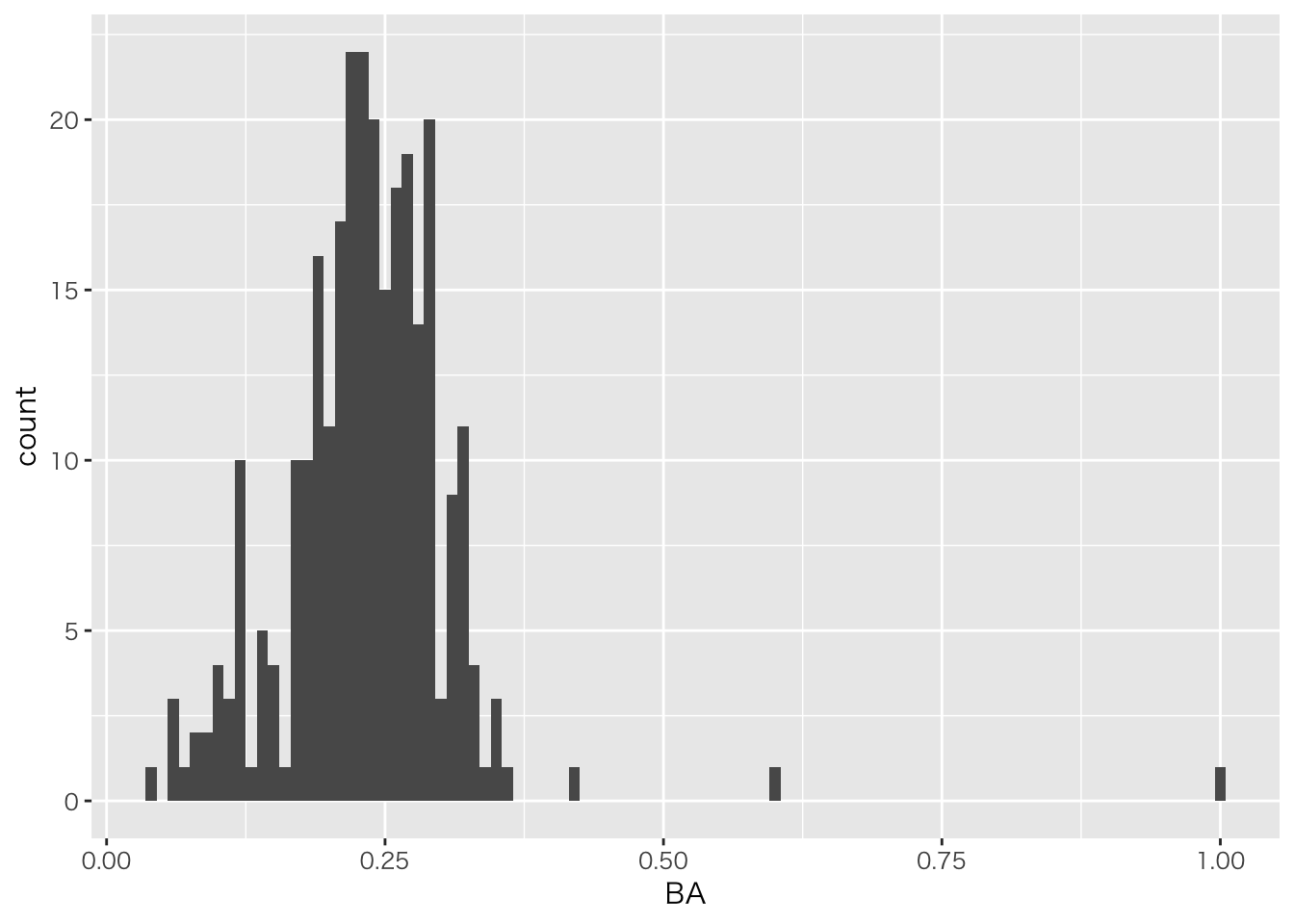
線形モデルの例。
# 書き方に注意
result.brms8 <- brm(HIT|trials(AB)~weight,data=baseball_dat7,family="binomial")## Compiling the C++ model## Start sampling##
## SAMPLING FOR MODEL '1954cda67d3d56cc4af3c182cf785a1e' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 0.000201 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 2.01 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 1.58861 seconds (Warm-up)
## Chain 1: 1.3675 seconds (Sampling)
## Chain 1: 2.95611 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL '1954cda67d3d56cc4af3c182cf785a1e' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 0.00017 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 1.7 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 1.87393 seconds (Warm-up)
## Chain 2: 1.31273 seconds (Sampling)
## Chain 2: 3.18667 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL '1954cda67d3d56cc4af3c182cf785a1e' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 0.000171 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 1.71 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 3: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 3: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 3: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 3: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 3: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 3: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 3: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 3: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 3: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 1.81627 seconds (Warm-up)
## Chain 3: 1.30983 seconds (Sampling)
## Chain 3: 3.12609 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL '1954cda67d3d56cc4af3c182cf785a1e' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 0.000169 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 1.69 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 4: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 4: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 4: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 4: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 4: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 4: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 4: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 4: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 4: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 1.70538 seconds (Warm-up)
## Chain 4: 1.21192 seconds (Sampling)
## Chain 4: 2.9173 seconds (Total)
## Chain 4:result.brms8## Family: binomial
## Links: mu = logit
## Formula: HIT | trials(AB) ~ weight
## Data: baseball_dat7 (Number of observations: 286)
## Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup samples = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
## Intercept -1.28 0.08 -1.43 -1.13 3292 1.00
## weight 0.00 0.00 0.00 0.00 3819 1.00
##
## Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample
## is a crude measure of effective sample size, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).pp_check(result.brms8)## Using 10 posterior samples for ppc type 'dens_overlay' by default.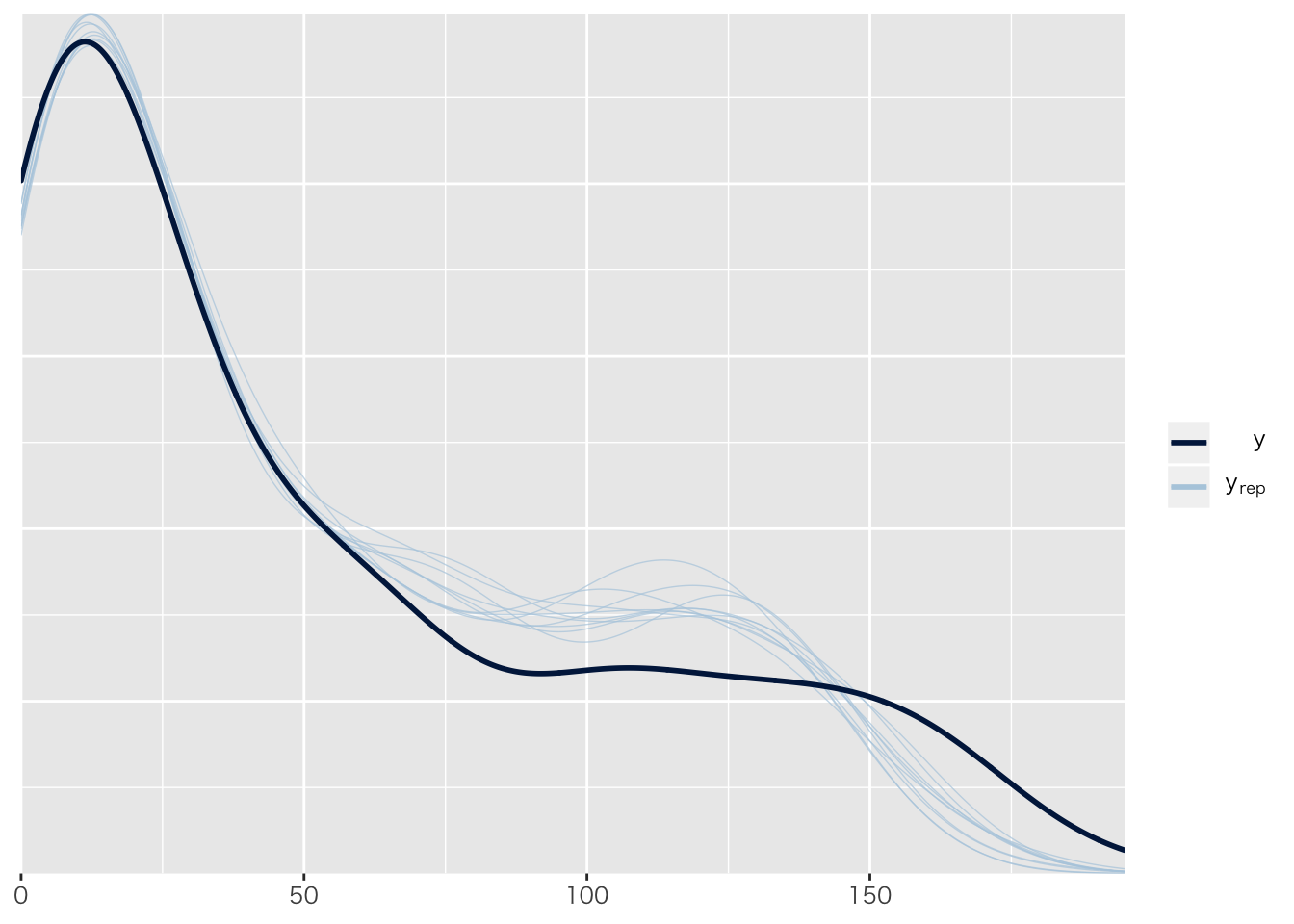
ロジスティック回帰
0/1データが従属変数の時は,特にロジスティック回帰という。
read_csv('baseball2019.csv') %>%
# 野手のデータ
dplyr::filter(position!="投手") %>%
dplyr::select(throw.by,weight) %>%
# 0/1データにする
dplyr::mutate(throw.01=ifelse(throw.by=="右",1,0)) -> baseball_dat8## Parsed with column specification:
## cols(
## .default = col_double(),
## Name = col_character(),
## team = col_character(),
## position = col_character(),
## bloodType = col_character(),
## throw.by = col_character(),
## batting.by = col_character(),
## birth.place = col_character(),
## birth.day = col_date(format = ""),
## 背番号 = col_character()
## )## See spec(...) for full column specifications.当てはめる回帰直線はロジスティックカーブを使う。
ggplot(data=data.frame(X=c(-10,10)), aes(x=X))+
stat_function(fun=function(x) 1/(1+exp(-x)))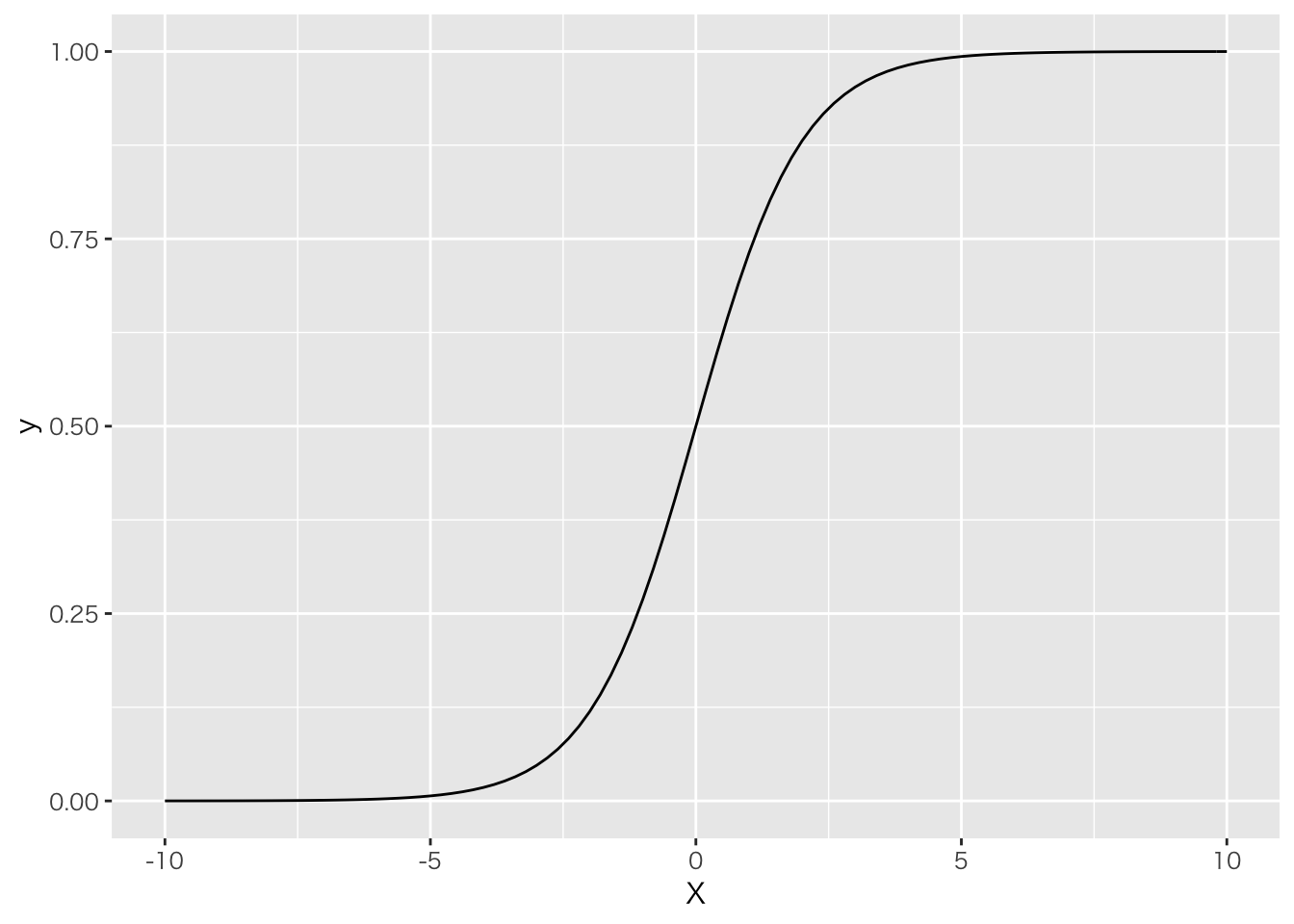
線形モデルの例。
result.brms9 <- brm(throw.01~weight,data=baseball_dat8,family="bernoulli")## Compiling the C++ model## Start sampling##
## SAMPLING FOR MODEL '7b5502e0422700fa03c91fff0d5e2f30' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 7.2e-05 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.72 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 0.214785 seconds (Warm-up)
## Chain 1: 0.167177 seconds (Sampling)
## Chain 1: 0.381962 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL '7b5502e0422700fa03c91fff0d5e2f30' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 4.4e-05 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.44 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 0.224253 seconds (Warm-up)
## Chain 2: 0.213649 seconds (Sampling)
## Chain 2: 0.437902 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL '7b5502e0422700fa03c91fff0d5e2f30' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 4.4e-05 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0.44 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 3: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 3: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 3: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 3: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 3: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 3: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 3: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 3: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 3: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 0.224216 seconds (Warm-up)
## Chain 3: 0.210684 seconds (Sampling)
## Chain 3: 0.4349 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL '7b5502e0422700fa03c91fff0d5e2f30' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 4.5e-05 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0.45 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 4: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 4: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 4: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 4: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 4: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 4: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 4: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 4: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 4: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 0.220199 seconds (Warm-up)
## Chain 4: 0.160407 seconds (Sampling)
## Chain 4: 0.380606 seconds (Total)
## Chain 4:result.brms9## Family: bernoulli
## Links: mu = logit
## Formula: throw.01 ~ weight
## Data: baseball_dat8 (Number of observations: 447)
## Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup samples = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
## Intercept 3.59 1.57 0.42 6.63 2942 1.00
## weight -0.01 0.02 -0.05 0.03 2997 1.00
##
## Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample
## is a crude measure of effective sample size, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).pp_check(result.brms9)## Using 10 posterior samples for ppc type 'dens_overlay' by default.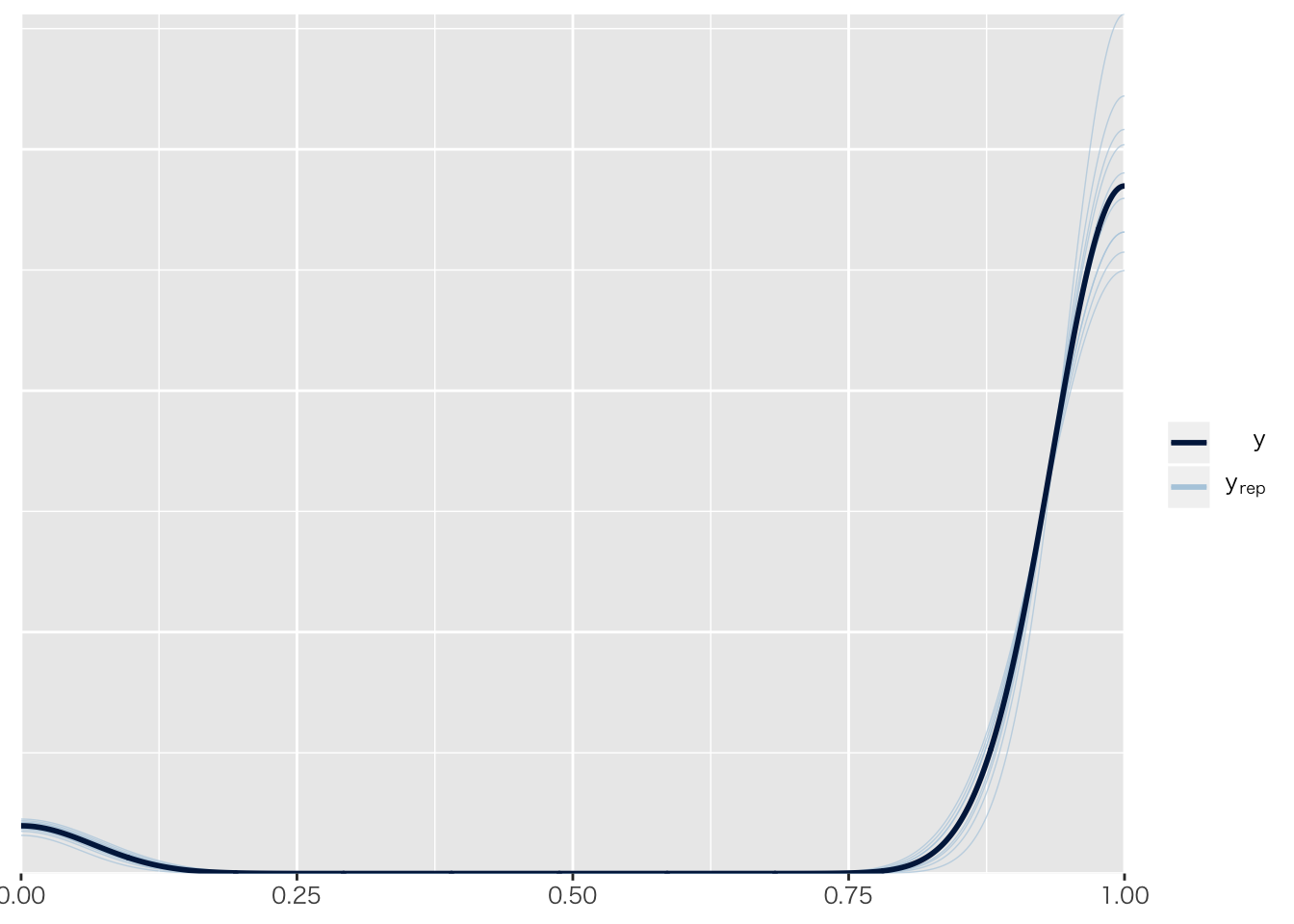
課題6
- 野球データの投手のデータを用いて,
- 勝利投手になった回数(変数名「勝利」)を契約年数(「years」)で予測する回帰分析と,
- 勝率(「試合」のうち,「勝利」の回数)を契約年数(「years」)予測する回帰分析を行うコードを書きなさい。
##
## SAMPLING FOR MODEL '84e3046c0af1517d6481f0c9f6abbda4' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 9.5e-05 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.95 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 0.29145 seconds (Warm-up)
## Chain 1: 0.277772 seconds (Sampling)
## Chain 1: 0.569222 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL '84e3046c0af1517d6481f0c9f6abbda4' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 6.1e-05 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.61 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 0.302591 seconds (Warm-up)
## Chain 2: 0.298087 seconds (Sampling)
## Chain 2: 0.600678 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL '84e3046c0af1517d6481f0c9f6abbda4' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 6.3e-05 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0.63 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 3: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 3: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 3: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 3: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 3: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 3: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 3: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 3: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 3: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 0.305621 seconds (Warm-up)
## Chain 3: 0.238853 seconds (Sampling)
## Chain 3: 0.544474 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL '84e3046c0af1517d6481f0c9f6abbda4' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 6.2e-05 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0.62 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 4: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 4: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 4: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 4: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 4: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 4: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 4: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 4: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 4: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 0.30971 seconds (Warm-up)
## Chain 4: 0.321397 seconds (Sampling)
## Chain 4: 0.631107 seconds (Total)
## Chain 4:## Family: poisson
## Links: mu = log
## Formula: win ~ years
## Data: baseball_dat9 (Number of observations: 337)
## Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup samples = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
## Intercept 0.90 0.06 0.78 1.01 3159 1.00
## years 0.00 0.01 -0.01 0.02 3683 1.00
##
## Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample
## is a crude measure of effective sample size, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).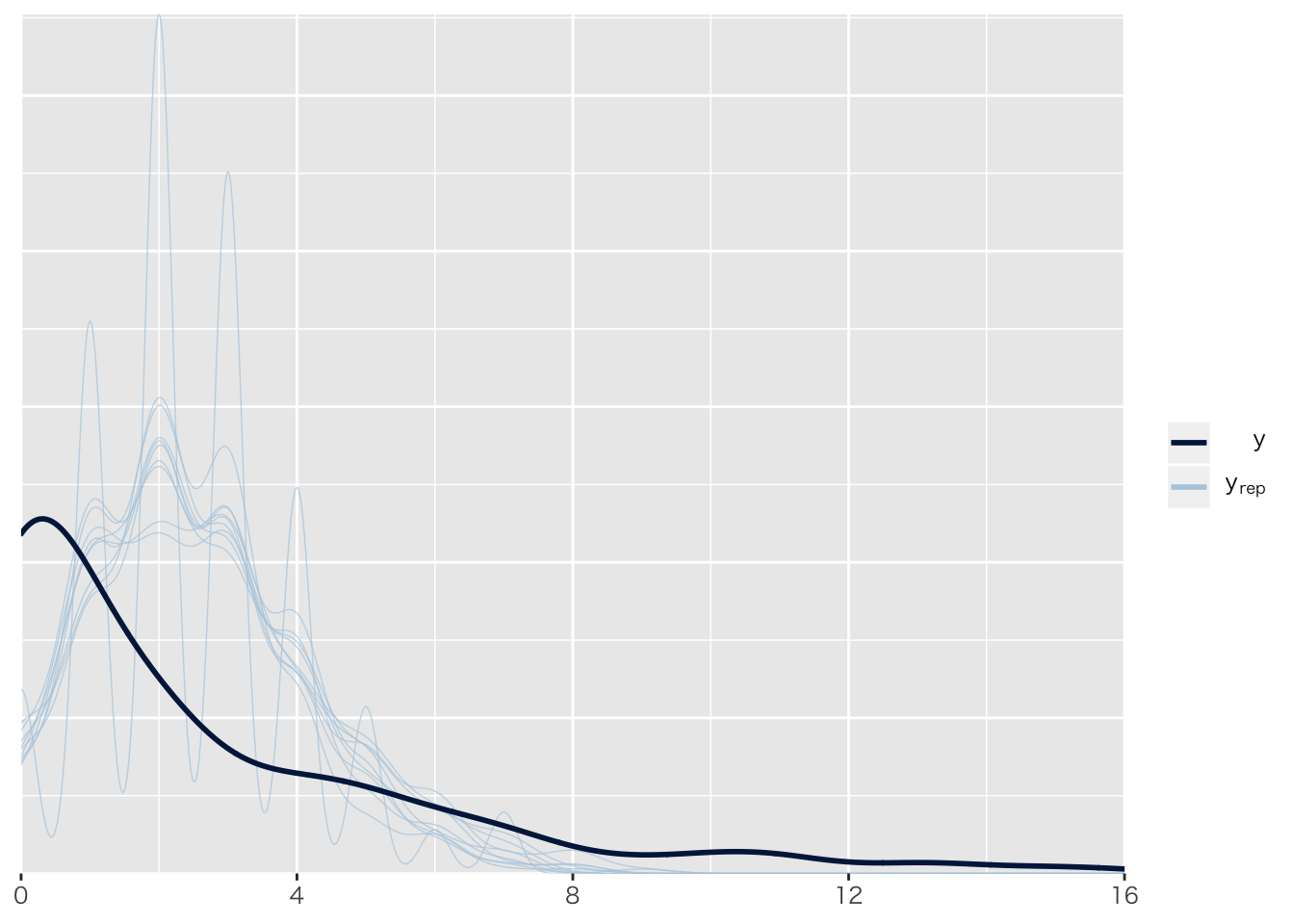
##
## SAMPLING FOR MODEL '1954cda67d3d56cc4af3c182cf785a1e' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 0.000229 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 2.29 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 0.991109 seconds (Warm-up)
## Chain 1: 1.17928 seconds (Sampling)
## Chain 1: 2.17039 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL '1954cda67d3d56cc4af3c182cf785a1e' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 0.000192 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 1.92 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 1.02118 seconds (Warm-up)
## Chain 2: 0.885995 seconds (Sampling)
## Chain 2: 1.90718 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL '1954cda67d3d56cc4af3c182cf785a1e' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 0.00022 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 2.2 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 3: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 3: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 3: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 3: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 3: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 3: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 3: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 3: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 3: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 0.956418 seconds (Warm-up)
## Chain 3: 0.856267 seconds (Sampling)
## Chain 3: 1.81269 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL '1954cda67d3d56cc4af3c182cf785a1e' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 0.000189 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 1.89 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 4: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 4: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 4: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 4: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 4: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 4: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 4: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 4: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 4: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 0.96961 seconds (Warm-up)
## Chain 4: 1.13957 seconds (Sampling)
## Chain 4: 2.10918 seconds (Total)
## Chain 4:## Family: binomial
## Links: mu = logit
## Formula: win | trials(Times) ~ years
## Data: baseball_dat9 (Number of observations: 337)
## Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup samples = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
## Intercept -1.88 0.06 -2.01 -1.75 3561 1.00
## years -0.02 0.01 -0.04 -0.00 4070 1.00
##
## Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample
## is a crude measure of effective sample size, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).